1. Background
Curcumin is a natural polyphenol compound with several biological and medicinal effects (Figure 1A) (1). Curcumin is derived from the curry spice turmeric that is produced in several Asian and South-American countries (1, 2). Accordingly, curcumin shows potential for the treatment of neurological diseases including Parkinson’s disease (PD) (1).
Monoamine oxidases A and B (MAO-A and MAO-B) are the two human isoenzymes of the monoamine oxidase enzyme (3). The MAOs are flavin-dependent mitochondrial enzymes that are involved in the oxidative degradation of various aromatic amine-containing neurotransmitters (3). Evidently, MAO-B is involved in neurodegenerative diseases including PD, which is related to dopaminergic neuron degeneration (4). Since MAO-B inhibitors can selectively inhibit the metabolism and increase the synthesis of dopamine (4), they are presently used as the support for the treatment of PD (3). For instance, safinamide is a highly selective and reversible MAO-B inhibitor that is part of the management of PD (Figure 1B) (5).
2. Objectives
Accordingly, we intended to evaluate the in silico interaction of curcumin with one subunit of the MAO-B enzyme in comparison with safinamide, the well-known MAO-B inhibitor.
3. Methods
The crystal structure of human MAO-B (PDB entry code 3PO7, 1.8A° resolution) in complex with zonisamide was selected from the Protein Data Bank (https://www.rcsb.org) (6). Chimera 1.8 software was used for editing the enzyme protein and building the extension file “pdb”. As the MAO-B forms a homo-dimer (7), we selected one subunit (chain B) for this docking procedure; moreover, all other molecules were deleted except FAD cofactor (8).
The structures of ligand molecules, including curcumin (CID: 969516) and safinamide (CID: 131682), were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov).
AutoDock Tools 1.5.6 software and AutoDock4 software were used for the docking procedure based on the previous studies (8, 9). Briefly, the grid of the enzyme was made by adjusting 68 × 68 × 72 grid points in xyz dimensions (8); moreover, the grid spacing was 0.375 A°, and the grid map was built based on the substrate-binding site (10, 11). All molecular docking simulations were based on the Lamarckian genetic algorithm (8). In addition, the other parameters, i.e. the population size: 150, the maximum number of energy evaluations: 2,500,000, and the maximum number of generations: 27000, were fixed (8). The output was an extension file “dlg” that contained the result of the docking procedure, including the binding energies (ΔG)s and inhibition constants (Ki)s of the conformations of each ligand (8, 12). Finally, the conformation of each ligand, presented the best down ΔG, was selected to display and discuss (12).
4. Results and Discussion
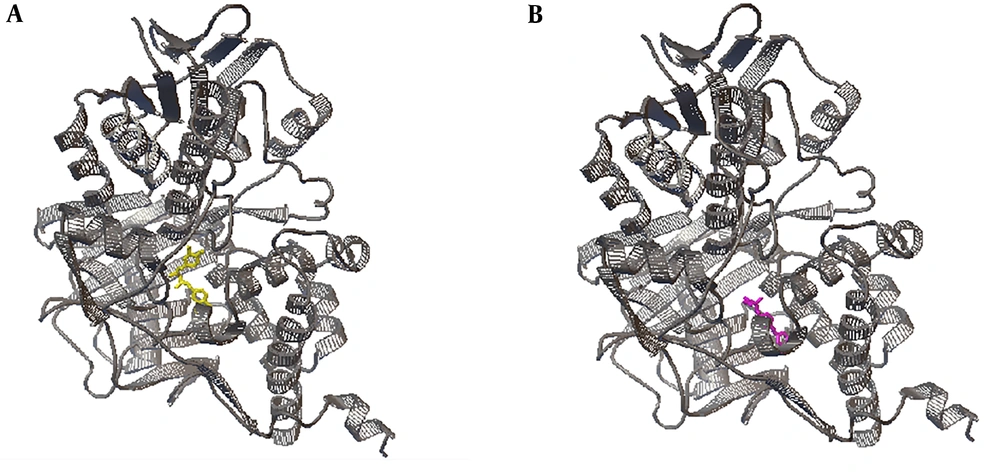
The results of the in silico interaction of curcumin and safinamide with one subunit of MAO-B are shown in Figure 2.
According to the docking results, the binding energies of the conformations of curcumin and safinamide, showing the best down ΔG, were -11.15 kcal/mol and -11.09 kcal/mol, respectively. As it has been considered that ΔG can determine the stability of any given protein-ligand complex (13), the near quantities of the ΔG(s) of curcumin and safinamide suggest that these ligands may form a nearly similar stability with the subunit of the MAO-B enzyme. Notably, the quantity of curcumin ΔG (-11.15 kcal/mol) is less than its binding free energy (-6.31 kcal/mol) calculated by a similar docking study (14). However, the different measures of these calculated binding free energies in two separate studies may be due to some differences in methods.
Moreover, the conformations of curcumin and safinamide, showing the best down ΔG, presented the Ki values of 6.72 nM and 7.48 nM, respectively. It has been mentioned that Ki can be theoretically used to rank order binding affinities of various inhibitors for a common enzyme (15). This in silico study revealed that the Ki values of curcumin and safinamide seem to be near mathematical measures; therefore, it could be proposed that curcumin and safinamide may present near binding affinities to MAO-B.
The results of this in silico study is in agreement with the MAO-B inhibitory effect of curcumin and its metabolite tetrahydrocurcumin in a mice model of PD (16); those experimental findings suggested that the inhibition of MAO-B by curcumin and tetrahydrocurcumin would be helpful in impeding the progression of PD (16).
In conclusion, the results of this in silico study revealed near quantities for ΔG and Ki of the conformations of curcumin and safinamide, showing the best down ΔG, in the interaction with one subunit of the MAO-B enzyme. Certainly, more experimental studies are required to disclose the similarity of curcumin with safinamide about the inhibitory effect on MAO-B for the treatment of PD.