1. Background
The medical application of silver nanoparticles (SNPs) has experienced a considerable growth during recent years in both diagnostic and therapeutic procedures. Due to their potent antimicrobial properties, silver nanoparticles are often utilized in the fabrication of textile and biomedical devices and in wound dressings as antimicrobial agents. Moreover, there are numerous papers reporting the empirical evidence about antibacterial, antifungal, and antiviral properties of SNPs. The considerable antimicrobial impact of SNPs on a large number of pathogens has made them a promising alternative to commercially available antibiotics (1).
During recent decades, the green synthesis of SNPs has been proposed as an appropriate, eco-friendly, quick, and cost-effective alternative to currently used chemical approaches. The biosynthesis of metal nanoparticles using bioactive compounds found in plant materials (enzymes, proteins, organic acids, and so forth) is environmentally safe and economically affordable (2). Among various species used so far for the bioreduction of metal nanoparticles, bacteria appear to be the most ideal platform for nanoparticle synthesis as they grow rapidly and produce a large biomass at lower costs (3). Some potential mechanisms have been proposed to explain this biological synthesis.
The most possible mechanism is the secretion of cellular reductases into the medium of growth by bacterial cells. Silver ions can be effectively reduced to silver nanoparticles by these enzymes. Moreover, it is possible to trap the metal ions by the functional groups that exist on the surface of bacterial cells. The entrapped ions can be further decreased by reductase enzymes, which in turn leads to the formation of nanoparticles. In fact, the biosynthesis of metal nanoparticles is a reduction process accelerated by the functional groups residing within the bacterial cells (4). Shewanella oneidensis is a bacterium with a high reducing power that efficiently reduces heavy metal ions and can be exploited as an appropriate cell factory for the production of nanoparticles (5).
It has been shown that biologically synthesized SNPs are powerful antimicrobial agents. Nonetheless, the antimicrobial effects of biologically synthesized SNPs at the molecular level are not properly known. Indeed, it is conspicuously evident in the relevant literature that most authors have only measured the inhibitory effect of SNPs on the growth of pathogens usually determined by calculating the minimum inhibitory concentration (MIC). Thus, it would be helpful to investigate if SNPs, especially at concentrations below MIC, can negatively affect the expression of virulence factors.
Escherichia coli is a Gram-negative and facultative anaerobic bacterium extensively used in biotechnology and medicine as a model organism. However, some pathogenic serotypes of E. coli are responsible for serious diseases in the human body, including different types of diarrhea, sepsis, urinary tract infections, and meningitis. Moreover, E. coli is also the most common facultative anaerobic bacteria residing in the gut flora of healthy individuals (6). Indeed, E. coli has been reported as a major cause of hospital infections around the world whose antimicrobial resistance may result in the failure of treating hospital-acquired infections (7).
Like other harmful bacteria, the pathogenesis of E. coli is mediated by so-called virulence factors including adhesins, toxins, and so forth (6). Hemolysin is a major toxin acting as a virulence factor in many pathogenic strains of E. coil. Alpha-hemolysin (Hly) is a well-known toxin produced by E. coli that damages erythrocytes via the formation of pores in their membranes (8). The clinical importance of alpha-hemolysin stems from the fact that the membrane pores formed by this virulence factor trigger the cascade events that finally result in the full hemolysis and degradation of the target cells (9).
2. Objectives
The present study was conducted to evaluate the efficacy of S. oneidensis in the green synthesis of SNPs as a fast and eco-friendly platform and to investigate the inhibitory effect of the synthesized SNPs on the growth of E. coli and the expression of Hly gene as a major virulence factor.
3. Methods
Silver nitrate stock solution was prepared by dissolving 3.39 g of AgNO3 in 100 mL sterile water to give a 200 mM solution. We harvested a healthy culture of S. oneidensis in the logarithmic stage and then centrifuged it at 5000 rpm for 5 minutes at 4°C. The supernatant was discarded and the biomass pellet was washed with sterile water three times to remove medium ingredients. The re-suspension of biomass was conducted in 47.5 mL of distilled water. Then, 2.5 mL of the 200 mM AgNO3 solution was added to get a final 10 mM concentration. A 50 mL suspension of S. oneidensis free of AgNO3 was used as the control (5).
The formation of SNPs was investigated by measuring the UV-Vis spectra of the solution in the range of 300 - 500 nm using a spectrophotometer device. The morphological properties of silver nanoparticles were investigated using transmission electron microscopy (TEM) by means of a Leo 912 AB high-resolution transmission electron microscope (HRTEM) that operated with an accelerating voltage of 120 kV (JEOL Inc, United States). A sample of the aqueous biomass solution was inserted in the carbon-coated copper grid and dried before microscopy. A sample of bioreduction solution was placed in a petri dish and oven dried for X-ray diffraction (XRD) assay. The dried sample was subjected to the XRD analysis by means of a Philips PW1480 X-ray diffractometer. The biosynthesized SNPs concentration was measured using a high-resolution ICP-OES spectrometer (SPECTRO ARCOS, Germany). For this purpose, a 50 mg sample containing both biosynthesized SNPs was diluted and filtered through filter paper and its final volume was adjusted to 25 mL. The concentration of SNPs was measured in this 25 mL sample (10).
The antimicrobial effect of SNPs was evaluated by a typical broth microdilution assay (11). E. coli was provided by Mashhad University of Medical Science, Iran. For the biologically synthesized SNPs, the minimum inhibitory concentration (MIC) was determined by the broth microdilution method in a 96-well standard ELISA plate. As the culture medium, Luria Bertani (LB) broth containing 105 CFU/mL of E. coli cells was employed. The final concentrations of SNPs were 0, 25, 50, and 100 μg/mL. No SNP was added to the negative control well. The minimum concentration of SNPs inhibiting the bacterial growth was assigned as MIC. To observe the growth dynamics of bacterial cells when exposed to SNPs, the growth curve was plotted at 0, 25, 50, and 100 μg/mL of SNPs by measuring optical density (600 nm) at varying time intervals.
The expression of alpha-hemolysin (Hly) under SNPs treatment was assessed by RT-PCR.
The RNA isolation and cDNA synthesis were implemented using the general procedure below (12). Primer pair sequences for RT-PCR of the virulence factor were 5’- TGAATCCTGTCGCTAATG-3’ as the forward primer and 5’- TATCATCCGACCTTTCACT-3’ as the reverse primer. The 16s RNA was used as housekeeping (reference) gene and internal control in RT-PCR assay. Forward and reverse sequences of the reference gene were 5’- CGTGCTACAATGGACAATACAAA-3’ and 5’- ATCTACGATTACTAGCGATTCCA-3’, respectively.
The expression of the virulence gene (Hly) was analyzed in a quantitative manner using a real-time PCR system (BioRad Inc, United States). The real-time PCR was conducted in a 20-μL reaction volume containing 0.5 μM of each primer and 10 μL of SYBR Green RT-PCR master mix (Genet Bio, South Korea). Quantitative real-time PCR experiments were carried out in duplicate for each sample. The real-time RT-PCR data were analyzed by the ΔΔCt method as described by Xiang et al. (12).
4. Results
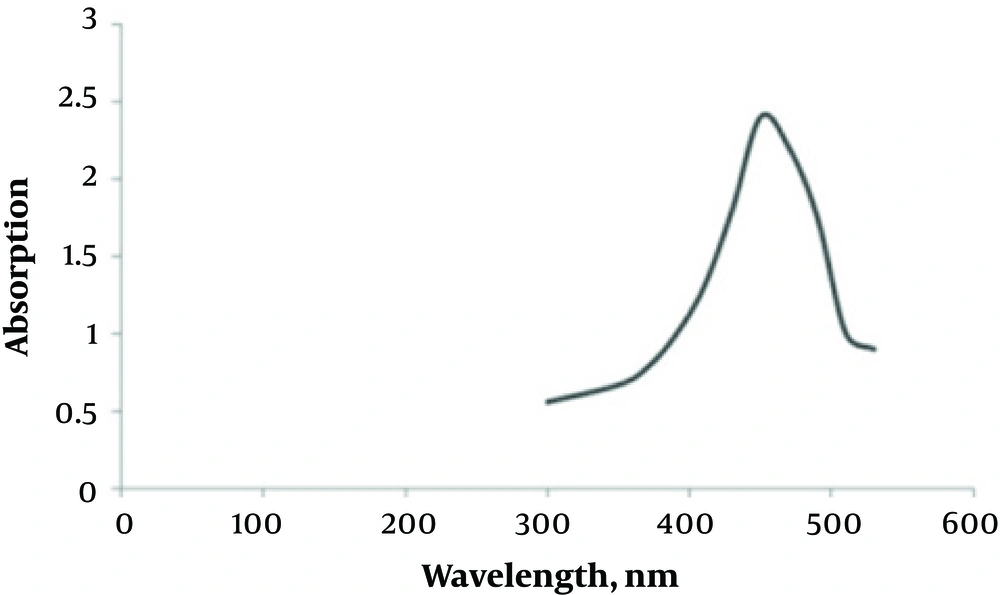
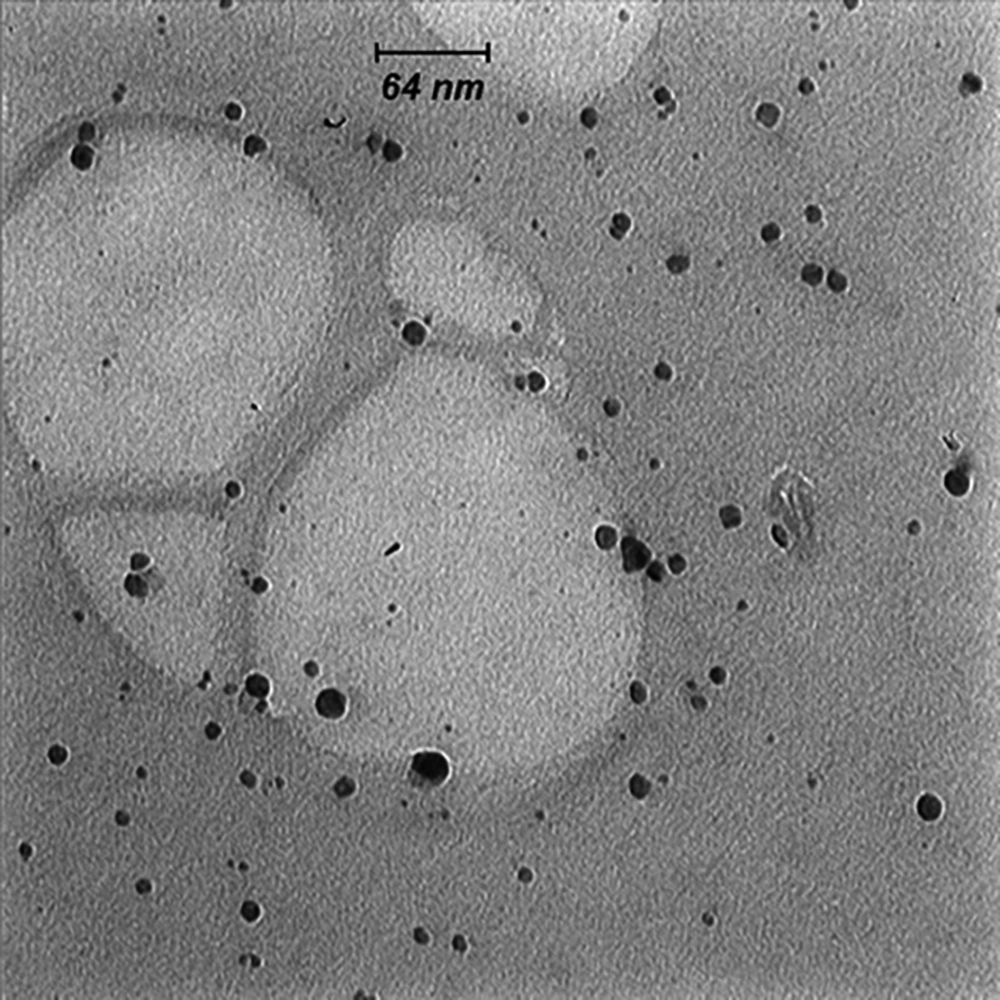
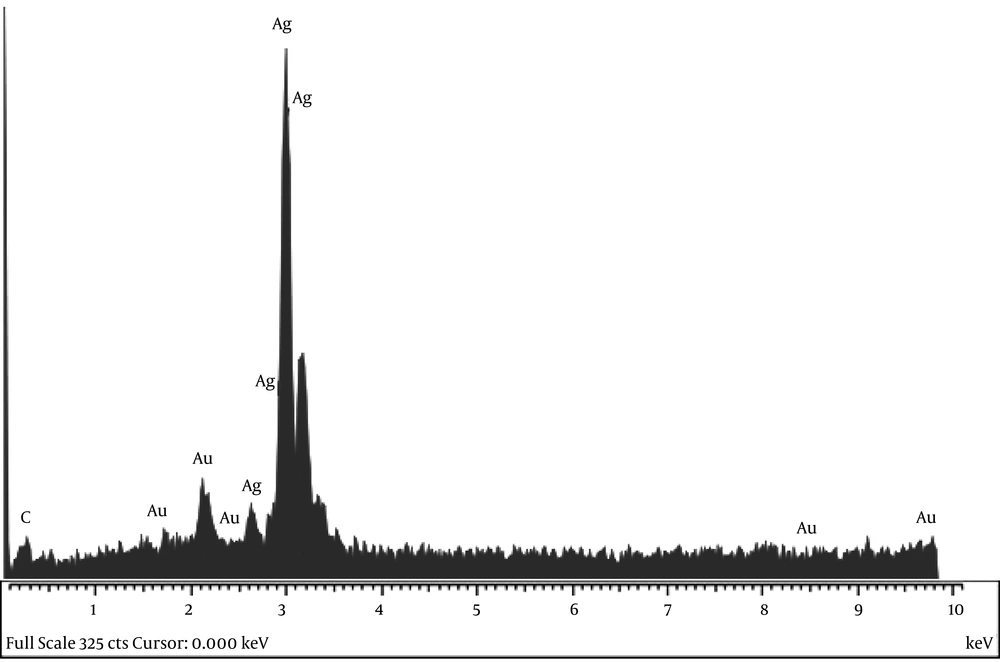
Adding silver nitrate to S. oneidensis suspension caused immediate color change. The UV-Vis spectroscopy revealed a surface plasmon resonance peak at about 450 nm, which confirmed the SNPs formation (Figure 1). The TEM microscopy was also used to demonstrate the morphology of biosynthesized SNPs (Figure 2). In general, TEM microscopy revealed that silver nanoparticles were of spherical shape with the size of about 10nm. Moreover, most of the SNPs had a circularity value of 1, confirming their spherical nature. The Energy Dispersive Spectrometry (EDS) revealed a sharp signal for Ag, which substantiated the silver NPs biosynthesis. The existence of silver nanoparticles at 3 keV was confirmed by the absorption peak (Figure 3).
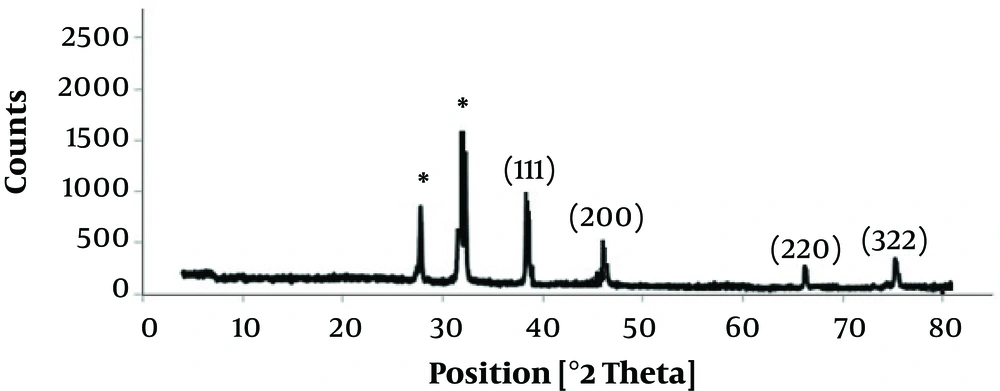
To characterize the diffraction properties and crystalline structure of the biosynthesized silver nanoparticles, the X-ray powder diffraction was used. The results of XRD indicated peaks equivalent to (111), (200), (220), and (322) Bragg reflections. This pattern revealed the existence of SNPs in the sample (Figure 4).
According to the XRD results, the particle size can be computed by Debye-Scherrer formula: d = kλ/βcosθ, where d indicates the nanoparticle size, k is Sherrer constant (0.9), λ is the X-ray wavelength (0.1541 nm), β indicates the full width at half maximum (FWHM), and θ is the diffraction angle. As suggested by the Scherrer equation, the mean crystallite size of the SNPs was in the range of 9 - 11 nm, which confirmed the particle size calculated by TEM images.
To determine the concentration of the biosynthesized SNPs, the ICP method was utilized. According to the results, SNPs concentration in a 25 mL sample that contained both bacterial biomass and SNPs was 2.934 mg/L.
The inhibitory effect of SNPs on the growth of E. coli was studied by serial microdilution technique. The results of the microdilution assay revealed that SNPs at the concentration of 50 μg/mL could hamper the pathogen growth. Therefore, the minimum inhibitory concentration (MIC) of SNPs was estimated at 50 μg/mL. The normal bacterial growth was found to be lower than the MIC value.
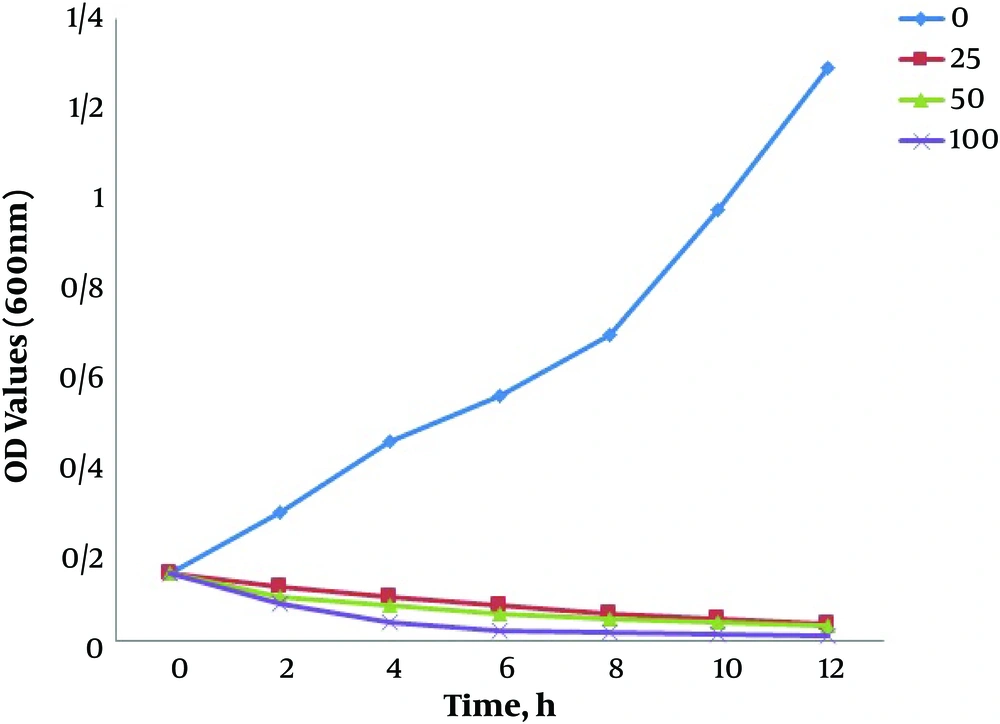
Growth kinetics of E. coli in a 6-h period was monitored under four concentrations of silver nanoparticles as 0 (control, non-treated), 25 μg/mL, 50 μg/mL (MIC), and 100 μg/mL (the highest concentration in this study) for further elucidation of SNPs effect on E. coli. Figure 5 shows the growth kinetics graph of the bacterium in these SNPs treatments. A typical growth pattern, which reached its highest level following 12 hours culture, was observed in the control group. The bacterial sample treated with 25 μg/mL and 50 μg/mL of SNPs followed a declining growth pattern so that after 10 hours, the growth level dropped to almost zero. The severity of growth was higher in the sample treated with 100 μg/mL of SNPs so that a growth decline was observed only after 4 hours, and the bacterial growth came to a complete halt 6 hours after the treatment.
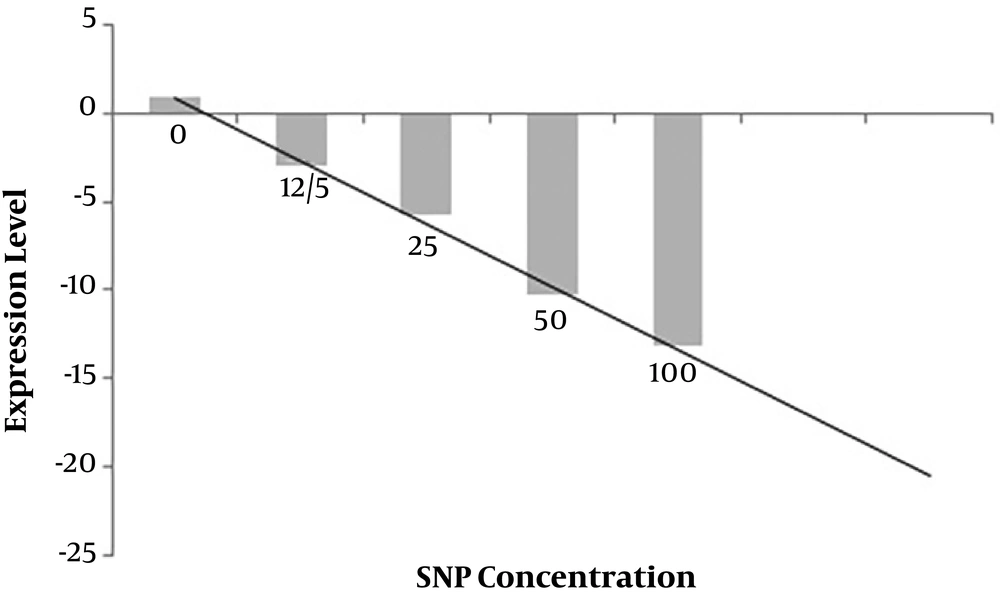
The influence of SNPs on alpha-hemolysin expression was evaluated via RT-PCR. RT-PCR results are depicted in Figure 6. The RT-PCR results revealed a dose-dependent reduction in the expression of alpha-hemolysin during treatment with varying concentrations of SNPs. The results suggested a fairly linear relationship between SNPs concentration and reduced alpha-hemolysin expression in the range of 0 to 100 μg/mL of SNPs.
5. Discussion
Nanobiotechnology has created a link between nanotechnology and biotechnology during recent years. As a branch of nanobiotechnology, the biosynthesis of metallic nanoparticle with extensive biological agents has several advantages over traditional mechanical and chemical synthesis procedures as noted earlier in this paper. Here, the biosynthesis of silver nanoparticles was investigated using a suspension of S. oneidensis. S. oneidensis is a natural bioremediator that accumulates and reduces huge metal and metallic pollutants. In this bioaccumulation process, non-toxic metals that contain compounds along with nanoparticles are produced from the trapped metal ions (13). As remarked by Suresh et al. (5), although numerous reports have been published on the bacterial-based biosynthesis of silver nanoparticles, all these methods produce either polydispersed or larger particles (> 20 nm).
The biosynthesis of silver nanoparticles was confirmed by the maximum peak at around 450 nm in UV-Vis spectroscopy. In other studies, a surface plasmon resonance peak in the range of 410 nm to 450 nm has been observed as an indicator of SNPs biosynthesis (10, 11). For example, Jioty et al. observed a surface plasmon resonance peak at 414 nm during the biosynthesis of SNPs using Urtica dioica leaf extract (14), which is in agreement with our results.
The TEM microscopy was used to analyze the morphology of the biosynthesized SNPs. Biologically synthesized nanoparticles can have different forms including rectangular, cubic, and spherical, in addition to other possible forms. In the present study, the TEM images revealed that the biosynthesized silver nanoparticles had a spherical shape with a mean diameter of 10.95 nm. The formation of spherical SNPs through green synthesis has been reported by other authors, as well. Korbekandi et al. reported the production of spherical SNPs using Althaea officinalis hydroalcoholic extract (15). In another study aiming at the biosynthesis of SNPs using Coffea arabica seed extract, the authors observed the formation of 20 – 30 nm spherical nanoparticles (16).
The EDS graph and XRD analysis confirmed the existence of SNPs in S. oneidensis suspension. In the EDS study, an absorption peak at 3 keV revealed the existence of silver nanoparticles in the solution. The bacterial biomass that contained silver nanoparticles was dried and then powdered for the purpose of XRD analysis. Accordingly, four peaks comparable to (111), (200), (220), and (322) Bragg reflections were found in this analysis. The XRD pattern achieved in this study was in agreement with previously determined Bragg reflections related to silver nanoparticles (17, 18). The particles size determined by Debye-Scherrer formula and RD data showed that the biosynthesized SNPs are 9 - 11 nm in average, which is consistent with the TEM results.
Following the characterization of SNPs, the serial microdilution technique was employed to study the antimicrobial effect on E. coli. The results of in vitro microdilution test indicated that SNPs could hamper pathogen growth at a concentration of 50 μg/mL.
The antimicrobial effects of silver nanoparticles have been widely reported in the literature (11, 19, 20). In this study, silver nanoparticles had a size of about 10 nm, making them ideal for the inhibitory effects on bacterial cells.
The nanoparticle size plays an important role in their inhibition of the microbial growth (21). It has been posited that nanoparticles of smaller sizes induce the enhanced antimicrobial effect due to their larger surface area and greater interaction compared to bigger particles (19). The bacterial growth inhibition caused by the effect of silver nanoparticles can appear in a variety of ways. For instance, silver nanoparticles interfere with sulfur found in biomolecules residing on the bacterial membrane or attack the respiratory chain and bacterial genome. In the end, these interferences can result in the bacterial cell death (22).
Besides exploring the effect of the SNPs on the growth inhibition of E. coli, we examined their impact on the expression of alpha-hemolysin as a crucial virulence factor of the pathogen. The results suggested that SNPs, even at concentrations of lower than the MIC value, could decrease the expression level of alpha-hemolysin. The inhibitory effect of SNPs on virulence factors of pathogenic bacteria has been reported by other authors, as well. For example, it has been recently reported that silver nanoparticles produced by biological systems are able to inhibit the expression of virulence factors of multidrug-resistant Pseudomonas aeruginosa strains (23). Alpha-hemolysin has been shown to be vital to the pathogenesis of lethal pneumonia in a murine model, leading to the extensive alveolar injury and associated epithelial barrier disruption (24). The adverse effect of the biosynthesized SNPs on the expression of virulence factors can medically contribute to the development of new antimicrobial medicines. Indeed, the neutralization of key virulence factors is a well-established strategy to identify effective preventative and therapeutic agents to fight bacterial infection (25).