1. Background
Mucopolysaccharidosis (MPS) is a set of inborn metabolic errors caused by an absence of specific lysosomal enzymes involved in glycosaminoglycan (GAG) catabolism (1). This metabolic block resulted in the GAGs accumulation in various organs, leading to progressive and multisystemic features (2). Based on the impairment of 11 involved enzymes, nine various clinical types and numerous subtypes of MPS have been identified (3). The subdivisions of MPS include Hurler Disease (MPS I), Hunter Syndrome (MPS II), Sanfilippo Syndrome (MPS III), Morquio Syndrome (MPS IV), Maroteaux Lamy Syndrome (MPS VI), Sly Syndrome (MPS VII), and hyaluronidase deficiency (MPS IX) (4).
It is very important to develop the genotypic catalog of specific disorders for a population. In this respect, there is inadequate research based on a limited number of cases in Iran. Therefore, the present study aimed to measure the rate of mutation and compare the data to the studies conducted in neighboring countries so as to identify the status of MPS disease in the Iranian population. Despite important information provided by genetic testing, there are limitations in this regard. Identification of common mutations prevents the need for next generation sequencing (NGS) based tests in MPS candidate patients. This results in lower diagnostic costs for patients and can be identified and categorized at a very low cost, and appropriate therapies can be considered.
2. Objectives
In this study, we examined the clinical and biochemical features and genetic analysis of 19 Iranian patients with MPS to identify novel MPS-associated mutations.
3. Methods
3.1. Ethical Statement
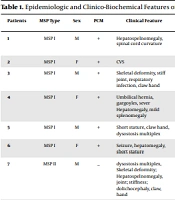
The sample included a total of 19MPS patients referred to the Taban Health Care and Diabetes Clinic (THCDC) from 2009 to 2015. The present study was approved by the Ethical Committee of National Institute of Genetic Engineering and Biotechnology (NIGEB) (IR.NIGEB.EC.1397.8.23.C), Tehran, Iran. Informed consent was obtained from patients. The clinical characteristics of MPS patients were summarized in Table 1.
| Patients | MSP Type | Sex | PCM | Clinical Feature | Main Symptom | Face Abnormality | Biochemical Urine GAG Accumulation Detected | MR | Cardiac Manifestations | Deafness |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MSP I | M | + | Hepatospelnomegaly, spinal cord curvature | Sever NDD, cloudy cornea, hearing loss | Coarse face, irregular’s dental, gum hyperplasia, corneal opacity | DS>HS | + | MI AI | BT |
| 2 | MSP I | F | + | CVS | # | # | # | # | # | # |
| 3 | MSP I | M | + | Skeletal deformity, stiff joint, respiratory infection, claw hand | Deafness, cloudy cornea, dolichocephalic, CHD, moderate NDD | gum hyperplasia, cloudy cornea Coarse face, | DS>HS | + | MI AS | Mixed |
| 4 | MSP I | F | + | Umbilical hernia, gargoyles, sever Hepatomegaly, mild splenomegaly | cloudy cornea 1 year, deafness 2years | Mild coarse face, irregular’s dental | DS, HS | + | MS | BT |
| 5 | MSP I | M | + | Short stature, claw hand, dysostosis multiplex | Macrocephaly, viceromegaly, | cloudy cornea, Coarse face, | DS>HS | + | MI | Mixed |
| 6 | MSP I | F | + | Seizure, hepatomegaly, short stature | Macrocephal, ombelical hernia | Facial dysmorphic features, mild Coarse face | DS, HS | N | MI AI | P |
| 7 | MSP II | M | _ | dysostosis multiplex, Skeletal deformity; Hepatospelnomegaly, joint; stiffness; dolichocephaly, claw, hand | Carpal tunnel, large tongue, Hydrocephaly, repeated diarrhea, large; tongue, J-shape sella; turcica | Coarse face, clear cornea, hypertelorism | DS, HS | + | Cardiomyopathy, Valvulopathy | BT |
| 8 | MSP II | M | _ | AF | # | # | # | # | # | # |
| 9 | MSP IIIB | M | + | Mild speech difficulty, insomnia, mild Skeletal deformity, | Psychotic, ADHD, NDR, hairsotysm, mild speech difficulty | Moderate Coarse face | HD CS | + | N | N |
| 10 | MSP IIIB | F | + | mild Skeletal deformity, | Difficult mood | Coarse face | ND | + | Valvulopathy | N |
| 11 | MSP IIIA | F | + | Mild speech difficulty | Aggressive behavior | Mild coarse face | HS CS | + | N | MILD BT |
| 12 | MSP IIIB | M | + | insomnia, mild Skeletal deformity, | ADHD, hairsotysm | Hair sot face | HS | + | N | N |
| 13 | MSP IIIB | F | + | moderate speech difficulty, mild short stature, Seizure, | Hairsotysm, NDR, otitis | Mild coarse face | HS | Mild + | N | N |
| 14 | MSP IIIA | M | + | Seizure, mild short stature | Claw hand, NDD, recurrent media otitis | MPS like phenotype | HS DS CS | + | Cardiomyopathy | Mixed |
| 15 | MSP IIIB | M | + | Mild speech difficulty, insomnia | ADHD, NDR, | Mild coarse face | HS, CS | + | N | N |
| 16 | MSP IIIA | M | + | Otitis media, speech difficulty, | NDD, NDR, Claw, hand, Psychotic | coarse face | Hs cs | + | N | Mixed |
| 17 | MSP IIIA | M | + | Seizure, Speech delay | Claw hand, Hairsotysm | Mild coarse face | ND | + | Valvulopathy | MILD BT |
| 18 | MSP IIIA | F | + | mild Skeletal deformity | NDR, Aggressive behavior | MPS like phenotype | HS CS | + | N | N |
| 19 | MSP IIIA | F | + | Insomnia, Skeletal deformity, | ADHD, Psychotic | coarse face | ND | Mild+ | CHD | Mixed |
3.2. DNA Extraction and Polymerase Chain Reaction (PCR)
Genomic DNA was extracted using PrimePrep Genomic DNA extraction kit (GeNet Bio) from peripheral blood samples of MPS patients, collected in tubes containing ethylenediaminetetraacetic acid (EDTA) in a final volume of 2 mL. Briefly, the PCR reaction was carried out in a total volume of 25 µL containing 50 ng of DNA, 2.5 µL of 10 × PCR buffer, 0.1 mM of each dNTP, 1 mM of MgCl2, 0.1 µM of each primer, and 0.3 units of Taq polymerase enzyme (CinnaGen, Iran) using a thermocycler (Eppendorf, Hamburg). The DNA was denatured at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min. The annealing temperature was based on the temperature (°C) value of each primer set (Table 2). The extension temperature was at 72°C for 1 min, with final extension at 72°C for 10 min.
| Primers | Sequences | |
|---|---|---|
| IDUA | Exon 1 | F-GAGTCATCGGTCCTCAGAGC |
| R-AGGACCCACCCACAAACAC | ||
| Exon 2 | F-CGCTGCCAGCCATGCTGAGGCTCG | |
| R-CCTCCCATCTGTGCCTCTGTAAGG | ||
| Exons 3-4 | F-GGGTTATTTTCCAAGGGGAAG | |
| R-CCAACCTATCCCTTGTCACC | ||
| Exons 5-6 | F-ATGCAGACGCCCTTCATC | |
| R-CCTGCTCCAGGATGGAGA | ||
| Exons 7-8 | F-CCACGACGGTACCAACTTCT | |
| R-TCCCCTTGGTGAAGGAGTC | ||
| Exon 9 | F-CTGGGGACTCCTTCACCAAG | |
| R-CAGGTAGCGCGTGACGTA | ||
| Exon10 | F-GTAAGCCGGGGTTCCAGG | |
| R-CGGTCCTCAGGGTTCTCC | ||
| Exon 11 | F-TGTGGGTGGGAGGTGGAG | |
| R-GAGGGAAGGGCTGTGATGG | ||
| Exon 12 | F-ACAGTGTGTGGGGTGAAGG | |
| R-TTGCTGGTGCACGTGTGT | ||
| Exons 13-14 | F-CCTAGGGGACATGAGATGGA | |
| R-CTCCAGCTGGGTCCTCATC | ||
| IDS | Exon 1 | F- CTGTGTTGCGCAGTCTTCAT |
| R- ATGCAGGAAAGGACAGATGG | ||
| Exon 2 | F- CCATCTGTCCTTTCCTGCAT | |
| R- TAACAAGATGTCCCGCACAA | ||
| Exon 3 | F- GCTGTGGCGATGCTTACCTCTG | |
| R- AAGAGAACCCAGACTCTGGACA | ||
| Exon 4 | F- GGCTTAGGGACCAGGAAGTC | |
| R- AACAAGTAGCACCCACCAGC | ||
| Exon 5 | F- CCTGCCTGGAAAACAAGAAA | |
| R- GGCCTTGACCTCTAAATCCC | ||
| Exon 6 | F- ACGTGGGGAATGCTAGTGAG | |
| R- GTTGGGAGAGTCCTGATCCA | ||
| Exon 7 | F- GCTGTGACTCTGTGGGTGAA | |
| R- CCAGGTTAAAAATGGGGGTT | ||
| Exon 8 | F- CAGCCTGTCAAGAATGAGCA | |
| R- ACCCCCAAAGCCTATGATTC | ||
| Exon 9 | F- CATATGGAGCCCAGACAGGT | |
| R- GGAAGGGAGCACATCACATT | ||
| SGSH | Exon 1 | F- GAGACCAGAGAGCCGGAG |
| R- ATTGACCACGGGTGGGG | ||
| Exon 2 | F- CTCACTCCCAGTGCTGTTTC | |
| R- GGGAGACGTGGCAGAGG | ||
| Exon 3 | F- GAGAACAGGTGCGGCAG | |
| R- ACCTCCTGGGCTCTGGC | ||
| Exon 4 | F- GAAGGGAGCAGAAAGGGTTG | |
| R- ATCCCGCCGGAAGACTC | ||
| Exon 5 | F- CCGAGGGGCTTCCTGTG | |
| R- CAAGCTCGTAGGAGGCCAG | ||
| Exon 6 | F- GTTCTGGGCTAACCCATTTG | |
| R- GACCCTCACCCACATTATGC | ||
| Exon 7 | F- GTCTACACACACCACCCGC | |
| R- CCCGTCCCAGATCCACTC | ||
| Exon 8 | F- GAGGGCAGCTCCTGTGTG | |
| R- CATCTCCAGAAGCTGAGCAA | ||
| Exon 9 | F- CTGGTACAAGGACCTCCGTC | |
| R- AAGGACAACTGTGTCCCCTG | ||
| NAGLU | Exon 1a | F- CCCAAGGGAGTATCCTGGTA |
| R- TGGCAGCCACAGAAGTCG | ||
| Exon 1b | F- CTTGGACACCTACAGCCTGG | |
| R- AGGCTCTGAAAGGCAGAGTG | ||
| Exon 2 | F- GGTACCTGGTCTCAGCTCCA | |
| R- GAAAACACCTACGGTGGCTC | ||
| Exon 3 | F- CCAGCACAAAGAAGCAATGA | |
| R- ATCTATCACCGATTCTGCCC | ||
| Exon 4 | F- CTGCGTGTATCCTGGGAGAT | |
| R- GGAATAAAATTCCCTCTCTGAGC | ||
| Exon 5 | F- GTGAACACTATGGCGGCTTC | |
| R- GTCCCTCTGCCTACCCCTAC | ||
| Exon 6a | F- GCCCTCTGTTTCATCACTCC | |
| R- GGTGGGAGACCCCATACC | ||
| Exon 6b | F- TGGTCTATTCCCTCATGGCT | |
| R- GTCACTAGCCAGCACCTCGT | ||
| Exon 6c | F- CTATGAGGAGGCAAGAAGCG | |
| R- AGCGGGGGTAATATTTGAGG | ||
| Exon 6d | F- CGTTCTCAGCAAGCAGAGGT | |
| R- TATAGCCCTGAGTCCTCCCA |
3.3. Sequencing Analysis
The double-stranded DNA of PCR products from MPS patients was examined using an automated ABI sequencing machine (Applied Biosystems 3100, Kavosh Fanavaran Kawsar Company, Tehran, Iran). The DNA sequences were confirmed for any nucleotide variation and then analyzed using Finch TV software (http://www.geospiza.com/finchtv/).
3.4. In Silico Analysis of the Variants
The impact of novel mutations was verified through Human Gene Mutation Database (HGMD). In order to predict the functional effects of novel variations, the sequence changes were evaluated using in silico prediction algorithms SIFT (5), polyphen (6), and I-Mutant 2.0 (http://folding.biofold.org/i-mutant/i-mutant2.0.html).
4. Results
4.1. Epidemiological and Clinical Data
In this study, 19 unrelated Iranian MPS patients were affected by MPS I (31%), MPS II (10%), MPS IIIA (31%), and MPS IIIB (26%) (Table 1).
4.2. Sequence Analysis
All the coding exons and intronic boundary regions of the IDUA, IDS, SGSH, and NAGLU genes were assessed by polymerase chain reaction (PCR) in all patients. The list of primers used in this study was presented in Table 2. Direct sequencing of the PCR products showed 16 variants in these genes, among which two nonsense and three missense alterations had been reported in the disease databases previously and two variants were present in the local population database (http://www.iranome.ir) (Table 3).
| Patients | MPS Type | Gene | Exon/ (Intron) | Variant | Protein Change | Point Mutation Type | Zygosity | Clinical Significance |
|---|---|---|---|---|---|---|---|---|
| 1 | I | IDUA | 1 | c.32T>G | p.L11R | Missense | Hom | Polymorphism |
| 2, 3 | 1 | c.T99G | P.H33Q | Missense | Polymorphism | |||
| 4 | 1 | c.99T>C | p.H33H | Missense | Disease Causing | |||
| 5 | 9 | c.1205G>A | p.W402* | nonsense | Disease Causing | |||
| 6 | 1 | c.33G>T | normal | silent | Disease Causing | |||
| 7, 8 | II | IDS | 5 | c.514C>T | p.R172* | nonsense | Hemi | Disease Causing |
| 11 | IIIA | SGSH | 2 | c.150G>T | normal | Silent | Hom | |
| 14 | 3 | c.256A>G | P.N86D | Missense | Disease Causing | |||
| 16, 18 | 4 | c.364G>A | p.G122R | Missense | Disease Causing | |||
| 17 | 4 | c.456G>A | p.I152I | Missense | Disease Causing | |||
| 19 | 1 | c.74G>A | p.R24H | Missense | Likely Pathogenic | |||
| 9 | IIIB | NAGLU | 3 | c.607c>T | p.R203* | nonsense | Hom | Disease Causing |
| 12 | 1 | c.259G>C | p.A87P | Missense | Polymorphism | |||
| 10 | 3 | c.683G>A | p.R228Q | Missense | Disease Causing | |||
| 15, 13 | 6 | c.2209C>A | p.R737S | Missense | Polymorphism |
Except the mentioned variants, remaining variants were not found in the disease databases (Clin Var, OMIM, HGMD, and literature in PubMed till to Jul 29, 2019). Six variants were predicted as disease-causing by Mutation Taster.
4.3. Amino Acid Substitutions
Among the cases, six patients had single base-pair substitutions in IDUA gene, five of these variants were detected in exon 1, and a single base pair substitution (c.1205G>A) was detected in exon 9 of one sample. Of the five mutations identified in the IDUA gene, three mutations were the missense type leading to a change in the amino acid sequence, one mutation was the nonsens type that leads to creation of stop codon, and one mutation was the silent type.
We found a single base pair substitution (c.514C>T) in exon 5 of IDS gene in one sample. This mutation is of the nonsense type and leads to the formation stop codons. In addition, four single base-pair substitutions in NAGLU gene were detected in five samples, including a single base-pair substitution (c.2209C>A) in exon 6 of two samples, two single base-pair substitution (c.607c>T and c.683G>A) in exon 3 of two samples, and a single base-pair substitution (c.259G>C) in exon 1 of one sample. Of these four mutations, three were missense mutations, and one was nonsense mutation. Moreover, our data showed five single base-pair substitutions in SGSH gene in 6 samples, including a single base pair-substitution (c.74G>A) in exon 1 of one sample, a single base-pair substitution (c.456G>A) in exon 3 of one sample, and two single base-pair substitution (c.364G>A and c.456G>A) were found in two and one of our samples, respectively.
Of these five mutations, four were missense mutations, and one was silent mutation that does not alter the amino acid sequence (type of mutations was summarized in Table 3).
5. Discussion
MPS is a group of hereditary, rare, and incurable ‘lysosomal storage diseases’ (7). It is estimated that 1 in 25,000 newborn children will have some type of the MPS in the United States in 2013 (8). MPS demonstrates remarkable genotypic heterogeneity, explaining the association of genotype-phenotype variability (9, 10). The present study indicated clinical and molecular features of 19 patients from unrelated Iranian families, manifesting various biochemical and clinical characteristics of MPS disease. Additionally, MPS is promptly diagnosed through urine and blood biochemical analysis (8, 11). However, a definite diagnosis of different types of MPS requires a wide range of comprehensive biochemical and molecular genetic techniques (12). In this study, to evaluate mutations of IDUA, IDS, SGSH, and NAGLU genes, we sequenced 19 blood samples acquired from MPS patients using a Sanger sequencing method. In our study, 15 variants in these genes were reported in MPS patients, of which eight variants were novel. Out of 15 variants, 9 variants were disease-causing mutations. Of the changes in IDUA identified in this study, the novel change c.99T>C (p.H33H) was found in one patient, which was a disease causing one, and it had not been published previously. However, the other variant c.1205G>A (p.W402*) was disease causing, and it had been reported by Zanetti et al. in Italian population (13) and Atceken et al. in Turkish population (14). The IDUA gene contains 14 exons (15). It should be noted that in our study, 80% of IDUA mutations were found in exon 1, and only 10% were in exon 9. Chkioua et al. showed novel splice site mutation in intron 11 of IDUA gene in four MPS I patients from four families from northern Tunisia (16). Sánchez reported that 14% of the IDUA gene mutations in his 7-member population were in exons 5 and 9 (17). Chkioua et al. reported that exon 1 mutations were detected in 75% of the patients, and exon 9 mutations were detected in 25% of patients in eight families with MPS I (18). Therefore, our results are consistent with those of Chkioua et al. The novel nonsense change c.514C>T (p.R172*) in exon 5 of IDS gene was found in more than one patient (in 10% our sample size). This variant was predicted as disease causing. Chistiakov et al. analyzed 17 children with hunter syndrome and found exon 5 of IDS gene mutations in 27% of mutations (19).
Exon 5 of IDS gene mutation accounted for only 5.5% of the total mutations observed in the study by Zhang (20). SGSH gene contains 8 exons, and interestingly, all the novel mutations in this study were found in exons 1 to 4. Mutational analysis on 23 patients from the UK with Sanfilippo syndrome type A showed that 30% of total mutations were located in exons 1 to 4 (21).
Yassaee et al. showed that SGSH mutations were in exons 2 and 7 in 11 families (22). Of the 5 variants found in the SGSH gene, three variants (c.364G>A (p.G122R), c.456G>A (p. p.I152I), and c.74G>A (p. p.R24H)) were predicted as disease causing, and the last two variants were not reported in any population; however, c.364G>A had been published in 2000 by Beesley et al. (21). In the study of NAGLU gene variants, three variants including (c.607c>T (p.R203*), c.259G>C (p.A87P), and c.683G>A (p.R228Q)) were predicted as novel pathogenic variants. In addition, Yassaee et al. revealed that NAGLU mutations were located in exons 2, 5, and 6 (22); but our data demonstrated that NAGLU mutations were in exons 1, 3, and 6. Similar studies have been performed in different countries. For example, Bekri’s study described the clinical and molecular features of 13 Algerian MPS 1 patients by molecular study of the IDUA gene (23).
Moreover, in a study conducted in 2014, molecular studies were performed in seven Mexican MPS I patients, and p.W402X was listed as a common mutation (17). Vafiadaki et al. identified the primary genetic lesion in 57 unrelated Korean MPS II patients, and they found various types of mutation in 42 patients (24). In the study by Beesley et al., mutational analysis was performed on the sulphamidase gene from 23 patients in the UK. In the study by Beesley et al., mutational analysis was performed on the sulphamidase gene from 23 patients in the UK, and 13 different new mutations were found (21). Our results showed positive mutation spectrum of IDUA, IDS, SGSH, and NAGLU genes in the Iranian cohort. The high fraction of detected novel variants highlights the mentioned genes mutation heterogeneity. However, this heterogeneity generates challenges in the interpretation of genotype and phenotype correlation. Meanwhile, if these mutations are studied and proven in a larger sample population, they could help to identify and categorize the patients with MPS symptoms. Due to the MPS heterogeneity and the clinical picture complexity, whole exome sequencing (WES) is usually recommended to all clients as a first tire test in Iran, which is a time consuming and costly test. If we have Iranian common mutations in the mentioned exons of target genes, we can screen the patients with MPS symptoms and diagnose them earlier at a lower cost.