Introduction
The Mycobacterium tuberculosis complex comprises many species an sub-species that cause tuberculosis in a variety of mammalian hosts an includes Mycobacterium bovis the principle cause of tuberculosis in cattle [1]. An important disease of domesticated cattle that has a major economic and health impact throughout the world, in contrast to M. tuberculosis, which is largely host restricted to humans, M. bovis is primarily maintained in cattles, in particular, domesticated cattle, although the pathogen can frequently be recovered from other mammals, including humans [2].
Genomic fingerprinting or DNA analysis by RFLP is a standard method for typing of M. tuberculosis complex [3].
Recently strains of M. bovis identified in east Africa and name of this group is African 2 (AF2) clonal complex, these strains had four or more IS6110 copies in their genomes and represents over 70% of all cattle isolates from each of these east African countries. Most M. bovis isolates frequently have only 1 copies of IS6110 in a common 1.9 kb, Pvull fragments [1, 4, 5]. So far, typing of M. bovis strains has not performed by IS6110-RFLP method in Iran and all research on M. bovis has been limited to PGRS-RFLP (Polymorphic GC- rich Repetative Sequences Restriction length fragment polymorphism) and DR-RFLP (Direct repeat-Restriction length fragment polymorphism) methods.
The aim of this study was evaluation of genetic patterns of M. bovis strains by IS6110-RFLP method for the first time in Iran.
Materials and Methods
Bacterial strains: In this experimental study, 25 lymph nodes specimens (mediastinal mesenteric and retropharyngeal) from tuberculin-positive cattle were collected from somethe provincesof Iran (Tehran, Markazi, Hamedan, Mazandaran, Isfahan, Fars, Kermanshah, Qom, West Azerbaijan, East Azerbaijan) and then digested and decontaminated with 5 mL N-acetyl-L-cysteine/sodium hydroxide (5 g/L N-acetyl-L-cysteinein 3.5 M NaOH and 0.05 M sodium citrate) for 15 min the digested specimens were centrifuged and neutralized with HCL (0.1 N), the sediments cultured on to Lowenstein Jensen (LJ) medium supplemented with pyrovate and incubated at 37°C for 8 weeks [6].
Identification: For identification of the isolates as M. bovis performed acid-fast staining and standard biochemical methods including niacin accumulation, nitrate reductase, catalase activity in 22ºC and 68ºC and resistant to thiophen-2-carboxylic acid hydrazide [7].
DNA extraction: The genomic DNA for RFLP was extracted from the visible colonies according to the method described by chloroform-isoamyl alcohol [8] (all of chemical materials that used for decontamination, identification and DNA extraction made of the Merck company, Germany). The concentration of extracted DNA was determined by nano drop (Nanodrop, USA) (700-1400 ng/µL). Genomic DNA was digested over night at 37°C with the restriction endonuclease Pvull (Roche, Germany). Digestes were separated by electrophoresis for 24 h in 1% agarose gels (Roche, Germany) in 1x TBE buffer at 40 V [9].
Southern blotting: Briefly, the gels were depurinated, denatured, neutralized. The DNA fragments resolved by gel electrophoresis were transferred onto charged nylon membranes (Roche-Germany) by southern blotting [9].
Hybridization and detection: Blots were prehybridized for 2-4 h in a rotating hybridization oven (Memert-England) and were hybridized overnight at 65°C in the same solution containing IS6110 probe was labeled by digoxigenin. Then anti-digoxigenin antibody conjugated with alkaline phosphatase (Roche-Germany) was added to the membrane and the signals were detected on the membrane by adding of the substrate BCIP/NBT (Roche, Germany) to detection buffer [9].
Results
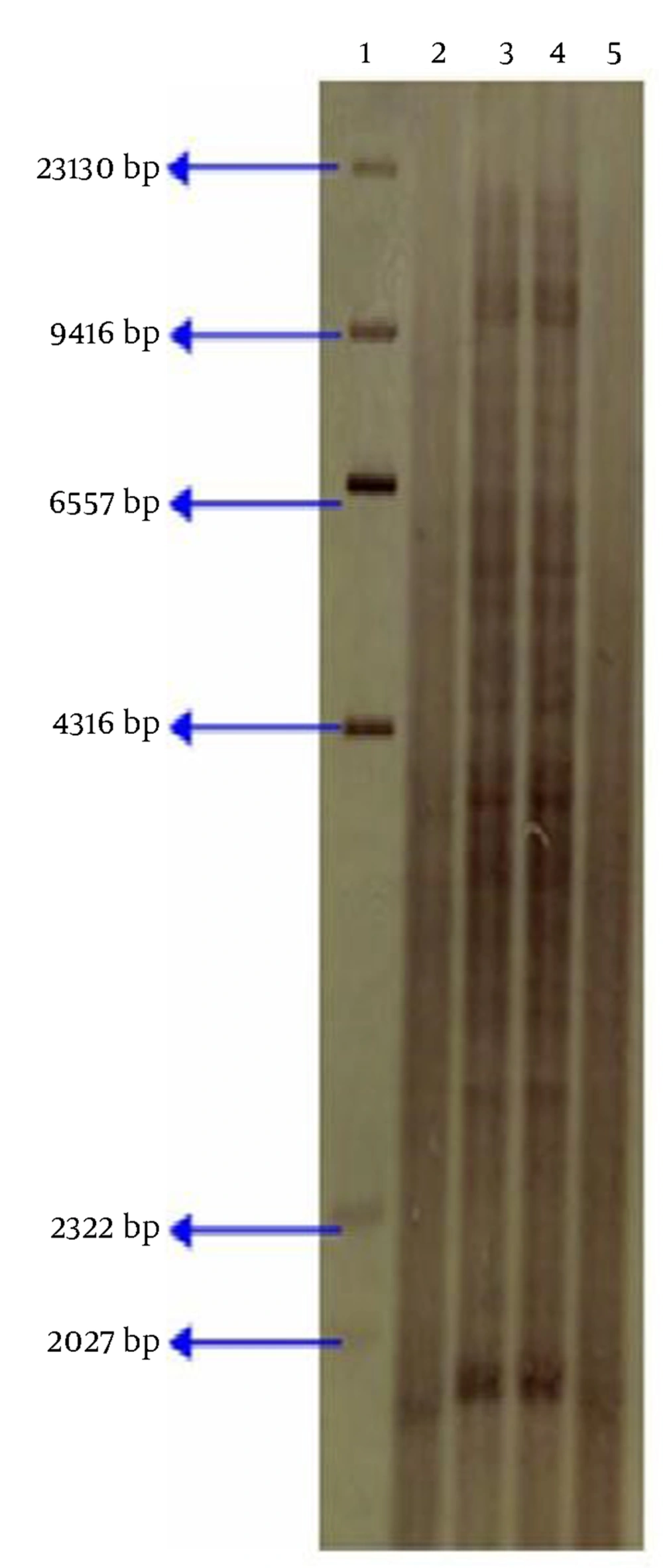
We collected 25 isolates of mycobacterium from lymph nodes of the tuberculin positive cattle and then all of them identified as M. bovis by acid fast staining and biochemical tests (Table 1). Twenty three isolates and standard strain of BCG (ATTC 1173P2) indicating presence of only one genomic copies of the IS6110 on 1.9 kb Pvull fragments by RFLP technique, and 2 isolates of M. bovis had multiple-band of IS6110 (with more than of 4 copies) (Fig. 1).
| Strains | Catalase 22°C | Catalase 68°C | Nitrate reductase | Resistant in TCH |
|---|---|---|---|---|
| - | - | - | - | |
| M. tuberclusis | + | - | + | + |
IS6110-RFLP patterns using PvuII lane 1: size marker Dig labeled (Roche, Germany), lane 2: M. bovis BCG (ATTC 1173P2) that showing only one copy of the IS6110, lane 3, 4: bovine M. bovis isolates with multiple-band of IS6110 from Iran, lane 5: bovine M. bovis isolate with only one copy of the IS6110 from
Compared and analyzed by special software, such as gel pro analyzer software (Media eybernetics, Italy) was done and it was found that the 2 strains are genetically similar. However in this study our results on IS6110 typing of M. bovis strains differ from previous results in Iran and observed strains of M. bovis with high copy number of IS6110 that isolated of cattle farm located in Mazandaran province, thus we report a first case of Bovine M. bovis isolates with high copy number of IS6110 by RFLP in Iran.
Discussion
In this research we observed 2 isolates of cattle farm located in Mazandaran province with high copy number of IS6110 that is a unique and specific case in Iran.
Today, the use of molecular typing techniques such as RFLP is useful to study genomic patterns. Molecular techniques used in this study have been selected according to world organization for animal health (OIE) recommendations [10]. Feizabadi et al. typed strains of M. bovis by IS6110-RFLP, but they did not observed more than 3 copies of IS6110 [11]. According to 1 copy of IS6110 in M. bovis isolated from cattle in the world, especially in Asia, the digested DNA hybridized with the IS6110 probe is skipped, on the other hand in the Spain 129 M. bovis strains from 5 different Spanish locations were fingerprinted using the IS6110 repetitive element and showed multiple copies (from 2 to 13) of IS6110 in a large proportion (47.4%) of the M. bovis strains isolated from cattle that accordance with our study [12, 13].
In other study in Sweden 45 isolates of M. bovis from humans and animals were analyzed by RFLP patterns probed by the insertion sequence IS6110. Most human isolates had patterns indicating the presence of only one or two genomic copies of the IS6110 insertion element, but isolates from M. bovis infections in 5 herds of farmed deer in Sweden showed a specific RFLP pattern with seven bands, indicating seven copies of the IS6110 sequence [14]. Recently researchers identified the strains of M. bovis with 4 or more copies of the insertion sequence IS6110 in east Africa, on the other hand the majority of M. bovis strains isolated from cattle, which are thought to carry only 1 or a few copies of IS6110, the results of this study are in agreement with our studies [4]. However M. bovis isolates bearing more than 4 copies of IS6110 in their genome have not been reported in Iran yet, thus we report a first case of M. bovis isolates with high copy number of IS6110-RFLP patterns in Iran that similar to AF2 clonal complex so it is likely that these strains (with high copy number of IS6110) of M. bovis entered in Iran and be involved in the epidemiology of Bovine tuberculosis in Iran; As in the previous study in Iran It was found there was a link between Iranian strains of M. bovis and those of its neighboring countries (i.e. Turkmenistan, Azerbaijan, Armenia, Turkey, Iraq, Pakistan and Afghanistan) [15]. Due to the lethality of tuberculosis and M. bovis transmission from animals and livestock products to human, we recommend that molecular analysis perform by IS6110-RFLP method for all of the strains of M. bovis in Iran and a molecular identification of the M. bovis strains available to be provided in accordance by IS6110-RFLP method.