1. Background
Cervical cancer remains a significant global health issue, ranking as the second leading cause of cancer-related deaths among women. This substantial burden is primarily attributable to its strong association with human papillomavirus (HPV), a prevalent and highly contagious virus. With over 90% of cervical cancer cases linked to HPV infections, there is a critical need for targeted prevention and management strategies (1). Effective strategies to mitigate the impact of cervical cancer necessitate a comprehensive understanding of the virus’s epidemiology, particularly the distribution and prevalence of its various genotypes (2).
Human papillomavirus is a small, non-enveloped virus with double-stranded DNA, encompassing nearly 200 genotypes classified as low-risk or high-risk based on their oncogenic potential. Low-risk HPVs (e.g., HPV6, HPV11) cause benign conditions such as genital warts, whereas high-risk HPVs (e.g., HPV16, HPV18) can induce squamous intraepithelial lesions that may progress to invasive squamous cell carcinoma if left untreated. Accurate genotypic identification is crucial for clinical and public health interventions (3, 4). This dichotomy underscores the critical need for precise genotypic identification to guide clinical and public health interventions effectively.
The epidemiology of HPV infection exhibits considerable regional variation, influenced by demographic, behavioral, and healthcare-related factors. These regional differences in HPV genotype distribution necessitate localized studies to generate pertinent data for effective public health strategies. For example, in regions with high prevalence rates of high-risk HPV types, intensified screening and vaccination programs may be warranted. Hence, understanding the distribution of HPV genotypes in specific populations is crucial for developing effective screening and vaccination programs tailored to local needs (5, 6).
2. Objectives
This study investigates the prevalence and genotypes of HPV in pap smear samples from women in Ahvaz, aiming to inform targeted interventions for reducing cervical cancer. The unique demographic of Ahvaz provides valuable insights into the distribution of HPV genotypes. The results will guide gynecologists in screening and tailoring strategies to the population’s specific needs. Findings will fill existing gaps in HPV genotype data for Ahvaz, aiding cervical cancer prognosis and supporting targeted public health interventions. Understanding the local distribution of HPV genotypes informs screening and vaccination decisions, ultimately reducing cervical cancer incidence and contributing to the global knowledge of HPV epidemiology.
3. Methods
3.1. Study Population and Sample Collection
3.1.1. Sample Size and Methodology
The study analyzed 290 pap smear samples from women attending private laboratories in Ahvaz, Iran (2017 - 2018) to investigate HPV infection. Patient data, including age and pap smear results, were collected. The sample size of 290 was determined based on the Morgan-Krejcie table, which provides a statistically significant sample size for a population of this scope, ensuring the reliability and validity of the conclusions drawn from the study. Samples were collected from Pasteur and Noor Clinics using a convenience sampling method due to logistical constraints and the availability of patients undergoing routine pap smear tests. While convenience sampling offers practical advantages, such as ease of data collection, it may introduce potential biases and limit the generalizability of the results to the broader population. It is important to consider these limitations when interpreting the findings of this study.
3.1.2. Age Range of Participants
The study included women across various age groups attending private laboratories for routine Pap smear tests. The participants were divided into the following age groups: 20 - 29 years (29%), 30 - 39 years (51%), 40 - 49 years (17.9%), and 50 - 60 years (2.1%). This diverse age range allowed for the assessment of HPV genotype prevalence across different age groups, facilitating a comprehensive understanding of the infection’s epidemiology in the region. The results indicated no significant correlation between the age of participants and HPV positivity (P-value > 0.05), highlighting the importance of targeted interventions irrespective of age.
The samples were collected from women attending these private laboratories for routine pap smear tests, ensuring that the study population was representative of individuals seeking healthcare services in these settings. However, since the sampling was not random, the results may not be fully representative of the general population of women in Ahvaz or other regions.
3.2. Human Papillomavirus Genome Extraction
The HPV DNA was extracted from the samples using the High Pure Viral Nucleic Acid Kit (Roche, Germany) in accordance with the manufacturer’s protocol. This step ensured the isolation of high-quality viral DNA suitable for subsequent PCR analysis.
3.3. PCR Testing for Human Papillomavirus Detection
The detection of HPV DNA was performed using conventional PCR with specific primers MY09/11 and GP5+/6+. Each reaction mixture contained 100 ng of DNA, with VP1-specific primers amplifying 450 bp and 150 bp fragments. A master mix was prepared for all samples, including controls. The samples were placed in microtubes containing the master mix and DNA and underwent thermal cycling using a Peqlab Peqstar thermocycler. Distilled water served as the negative control, while a confirmed positive sample was used as the positive control.
3.4. Primer Sequences Used in PCR Reactions
The primers used in the PCR reactions were as follows:
MY09: 5’-CGTCCMARRGGAWACTGATC-3’ (450bp)
MY11: 5’-GCMCAGGGWCATAAYAATGG-3’
GP5+: 5’-AAAAATAAACTGTAAATCATATTC-3’ (150bp)
GP6+: 5’-TTTGTTACTGTGGTAGATACTAC-3'
3.5. Human Papillomavirus Genotyping in Positive Samples
The HPV genotyping was conducted on samples that tested positive for HPV DNA using multiplex PCR with 20 pairs of primers, as referenced in a previous study. For each patient, four microtubes were prepared, with five pairs of primers added to each reaction mixture. The microtubes were then placed in a thermocycler and subjected to the specified thermal cycling program.
Following PCR amplification, the products were electrophoresed on a 2% agarose gel. The bands observed in each well were compared to standard size markers to determine the HPV genotypes. The sequences of the primers used for HPV genotyping were specified in the study. In all PCR assays, the beta-globin gene was used as an internal control to ensure the reliability of the results.
3.6. Data Analysis
Patient data were analyzed using SPSS version 19. Statistical evaluations were performed using chi-square tests and independent t-tests. Descriptive statistics were presented through tables, charts, means, and standard deviations. A significance level of P-value < 0.05 was considered for all statistical tests conducted in this study.
3.7. Ethical Considerations
The study was conducted at no cost to the participating patients. All expenses for laboratory tests and necessary materials were covered by the researcher. Patient confidentiality was maintained throughout the study, and no information was shared with any external entities or individuals.
4. Results
4.1. PCR Results
In this study, the PCR test was conducted using specific primers GP5+/6+ and MY09/11 to detect the HPV genome. Out of the 290 samples analyzed, 216 samples (74.5%) tested positive for the presence of the virus, while 74 samples (25.5%) were negative.
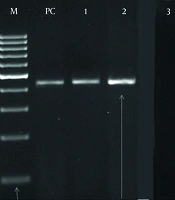
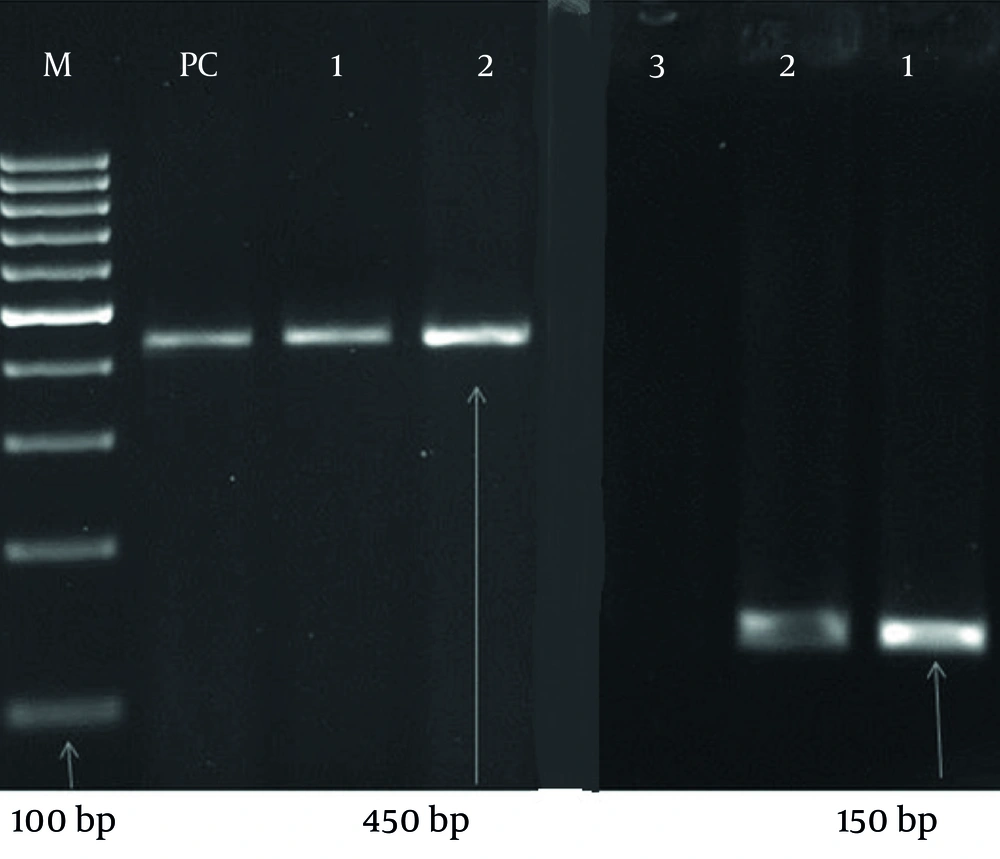
In the negative control well, no bands were observed, indicating the absence of contamination or false positives. Conversely, in the positive control well, bands corresponding to 450 bp and 180 bp were clearly visible, confirming the accuracy and reliability of the PCR assay (Figure 1).
PCR detection of the human papillomavirus (HPV) genome in 290 samples using specific primers GP5+/6+ and MY09/11. Among the samples, 216 (74.5%) tested positive, while 74 (25.5%) were negative. The negative control well showed no bands, indicating no contamination, whereas the positive control well showed bands at 450 bp and 180 bp, confirming the accuracy of the PCR assay.
4.2. Genotyping of Positive Samples
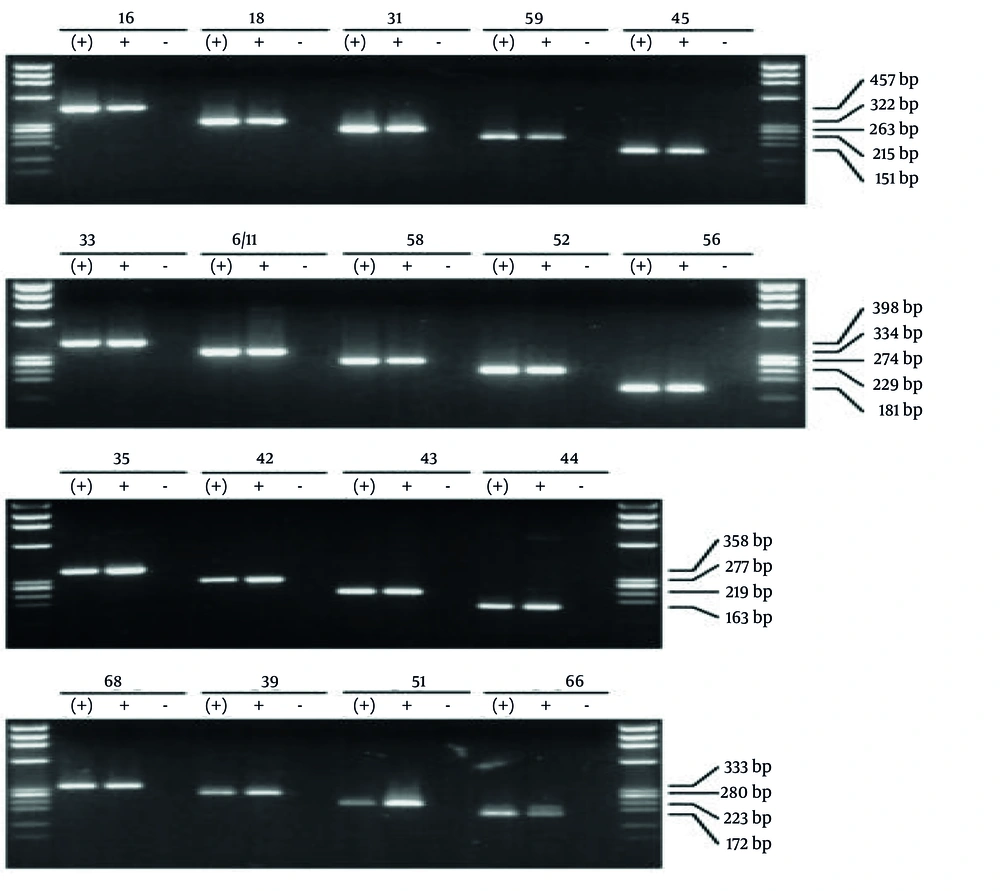
For the samples that tested positive for the HPV genome, genotyping was conducted using 20 pairs of specific primers through multiplex PCR. Each sample was processed in four wells, and the resulting bands were compared to standard size markers to ascertain the HPV genotype of each sample. The images depicting the standard markers used for comparison are presented in Figure 2.
The image illustrates the bands from reference samples used to determine the low-risk and high-risk human papillomavirus (HPV) genotypes. Genotyping was performed using the multiplex PCR method with 20 pairs of specific primers. Each sample was processed using four wells, and the obtained bands were compared with those of the reference samples shown in the figure to accurately identify the genotypes.
Among the total samples, 200 (68.9%) tested positive for low-risk genotypes, either as single infections or as co-infections with multiple genotypes. Additionally, 135 (46.6%) samples were positive for high-risk genotypes. Among the low-risk genotypes, HPV6 was the most prevalent, detected in 70.7% of the samples. This was followed by HPV11 (35.2%), HPV81/62 (20.7%), HPV43 (19.6%), HPV42 (12.1%), HPV54 (4.8%), HPV44 and HPV55 (3.4% each), HPV67 (1.7%), and HPV61 (0.7%) (Table 1).
Among the high-risk genotypes, HPV39 was the most frequently identified, accounting for 29.7% of the positive samples. This was followed by HPV51 (21.4%), HPV52 (18.9%), HPV66 (14.5%), HPV16 (12.7%), HPV18 and HPV45 (each 5.8%), HPV53 (5.5%), HPV31 (5.2%), HPV35 (3.8%), HPV82 (2.1%), and HPV56, HPV68, and HPV59 (each 1.7%) (Table 2).
| Genotype | Percentage |
|---|---|
| 39 | 29.7 |
| 51 | 21.4 |
| 52 | 18.9 |
| 66 | 14.5 |
| 16 | 12.7 |
| 18 and 45 | 5.8 |
| 53 | 5.5 |
| 31 | 5.2 |
| 35 | 3.8 |
| 82 | 2.1 |
| 56 and 68 | 1.7 |
| 59 | 1.4 |
Prevalence of High-risk Human Papillomavirus Genotypes in the Samples
4.3. Coinfection Analysis
Table 3 presents the percentage of samples that were positive for multiple HPV genotypes. The distribution is as follows.
| Description | Samples |
|---|---|
| Multiple HPV genotype infection | 216 (74.5) |
| Multiple HR genotype infection | 135 (46.6) |
| Multiple LR genotype infection | 200 (68.9) |
| Infection with both 16 and 18 genotypes | 9 (3.1) |
| Infection with both 6 and 11 genotypes | 83 (28.6) |
| Infection with 6, 11, 16 and 18 genotypes | 6 (2.1) |
4.4. Age Analysis and Genotype Correlation
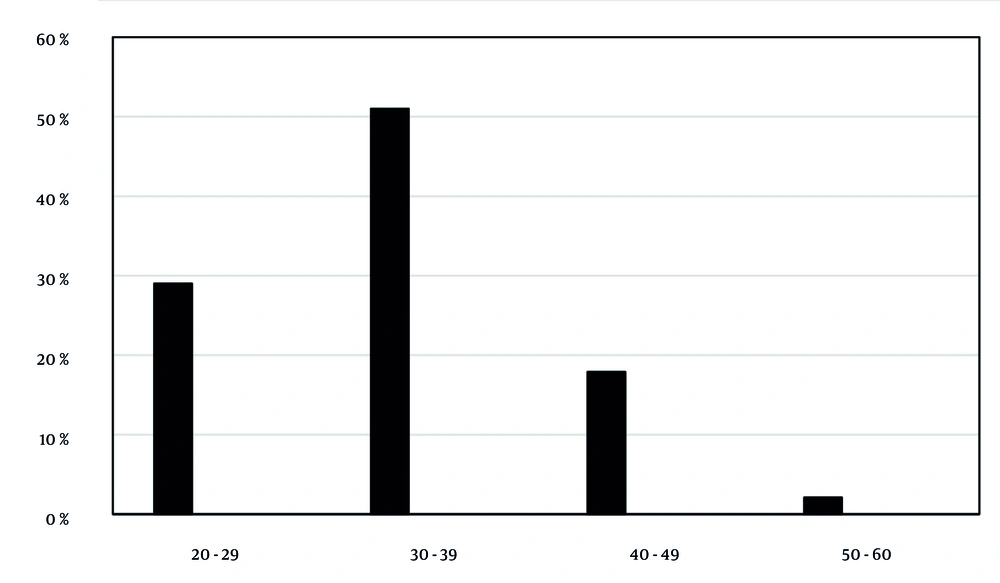
The patients' ages ranged from 20 to 60 years (average 38 years). They were categorized as follows: 84 patients (29%) aged 20 - 29, 148 patients (51%) aged 30 - 39, 52 patients (17.9%) aged 40 - 49, and 6 patients (2.1%) aged 50 - 60 (Figure 3). The 30 - 39 age group had the highest number of positive cases, but no significant correlation was found between patient age and HPV positivity.
Age distribution of patients and human papillomavirus (HPV) positive cases. Patients were divided into age groups: 20 - 29 years (29%), 30 - 39 years (51%), 40 - 49 years (17.9%), and 50 - 60 years (2.1%). The highest number of positive cases was in the 30 - 39 years group. No significant correlation between age and HPV genotype was found (P-value > 0.05).
4.5. Patient History Results
Patient histories from Ahvaz revealed that five individuals with genotypes 16 and 18 engaged in high-risk behavior with multiple sexual partners, while seven reported that their spouses had high-risk behaviors. Among the positive cases, 18 patients had a history of treated genital warts. Pap smear results showed that none of the women with a positive HPV genome had malignant or cancerous cells.
5. Discussion
Human Papillomaviruses are a group of small, non-enveloped viruses with double-stranded DNA that belong to the Papillomaviridae family (7). These viruses predominantly infect squamous epithelial cells in various anatomical locations. To date, approximately 200 types of HPVs have been identified. The nature of HPV infections was first recognized nearly 70 years ago, although warts were known in ancient Greece and Rome and were considered infectious (8). It was not until the early 20th century that the viral nature of human warts was confirmed when cell-free filtrates from lesions were found to be infectious (9). Due to host species specificity and the lack of suitable cell culture systems, studying the biology of HPVs has been challenging.
The HPVs cause benign skin lesions (warts) and mucosal lesions (condylomas). Certain types are linked to cervical cancer and other anogenital tumors. CRPV was the first identified DNA tumor virus. Cervical cancer is the second leading cause of cancer deaths among women. Effective prevention, early diagnosis, and timely treatment reduce cervical cancer mortality. Molecular methods like PCR are vital for the definitive and early diagnosis of HPV, especially due to limitations in traditional methods (10-14).
In 90% of cases, cervical cancer is caused by HPV infection (15). However, many individuals with HPV infections do not develop cervical cancer. Other risk factors include smoking, a weakened immune system, oral contraceptive use, early onset of sexual activity, and multiple sexual partners. Cervical cancer typically develops from precancerous changes over a period of 10 to 20 years (16). Various types of cervical cancer exist, with approximately 90% being squamous cell carcinoma, 10% adenocarcinoma, and a small number of other types. Diagnosis involves cervical screening followed by biopsy. Imaging is then used to determine the extent of the disease.
Analyzing the genomes of cancerous tissues provides valuable information for identifying diagnostic and preventive factors (17). Numerous risk factors associated with the pathogenesis of this disease have been identified, including family history, hormones, and smoking. While genetic factors play a role in cancer development, they account for a limited percentage of cervical cancer cases, indicating that environmental factors also contribute significantly (17).
5.1. Comparison of Study Results
In the current study, 74.5% of samples tested positive for HPV genome presence. This contrasts with findings from various Iranian studies: Yousefzadeh et al. (18) reported 31.1%, Sabat et al. (19) reported 48%, and Salehi-Vaziri et al. reported 45.4% (20). Internationally, McGill et al. in Grenadian women reported 33%, in Brazil 12.6%, Hernandez-Rosas in Mexican women 12%, and Stevens et al. in Melbourne 15.8%. Compared to other studies, the prevalence rate of HPV in the present study is higher, which may be due to the smaller sample size or the higher sensitivity and specificity of the methods used, as methodologies vary across studies (18, 21-25).
Among low-risk genotypes, genotype 6 had the highest prevalence at 70.7%, which aligns with the findings of many other studies. The prevalence of this genotype is not surprising, as it can also be found in the genital tract of healthy individuals. For high-risk genotypes, genotype 39 had the highest prevalence at 29.7%, consistent only with Jiang et al.'s study in China. Other studies reported different high-risk genotypes, suggesting a wide geographic distribution of various HR types in different regions of Iran and around the world (26).
The prevalence rates of HR types 18 and 16 in our study were moderate at 12.7% and 5.8%, respectively. This contrasts with lower rates reported by Yousefzadeh et al., Salehi-Vaziri et al., and Jiang et al. (18, 20, 26). In the studies by Nascimento, Kornal, and Jacob, these types were nearly absent, possibly due to differences in sample collection methods and the geographic distribution of these genotypes (27, 28). However, Cranston et al. reported a 93% prevalence among individuals with multiple sexual partners, with type 16 observed in 36% of cases (29).
The highest positive rates in our study were in the 30 - 39 age range, but no significant correlation between age and genotype type was observed. Stevens et al. noted a significant decrease in concurrent infections with multiple high-risk HPV genotypes with increasing age (21).
5.2. Conclusions
The study revealed a high prevalence of HPV detected via PCR, indicating both an effective methodology and a heightened regional risk. Most positive cases were low-risk; however, targeted education and preventive measures are needed to address high-risk genotypes. Expanding screening and preventive strategies can help reduce HPV-related diseases in the region.