1. Background
Schizophrenia is a severe mental illness that blurs the line between fantasy and reality, causing individuals to struggle with understanding and distinguishing what is real. This disorder greatly impairs a person's perception of reality. Schizophrenia leads to difficulties in personal, social, and occupational functioning, putting significant stress on the individual, their family, and caregivers. Patients struggling with this disorder often experience fear and display abnormal behaviors, making schizophrenia one of the most disabling mental illnesses (1).
Common symptoms of schizophrenia involve hallucinations, delusions, disrupted thought patterns, reduced ability to perceive reality, deterioration of personality, loss of appetite, feeling manipulated by external influences, and experiencing depression (2). Sophisticated brain imaging techniques have revealed distinct variations in the brains of individuals with schizophrenia when compared to those of healthy individuals. Some individuals with schizophrenia may exhibit underdeveloped areas in their brains compared to those who are healthy (3).
Various factors can contribute to the development of schizophrenia, including environmental influences like substance abuse, abnormalities in brain structure, genetic predisposition, and experiences of deprivation in childhood. Childhood deprivation or abuse is a significant factor in the onset of this disorder (4). This condition affects men and women equally, with a prevalence rate of one in every hundred individuals. Studies indicate that genetics contribute to approximately 50% of the development of schizophrenia (5, 6).
Based on recent data from the Our World in Data website, the prevalence of schizophrenia in both men and women in Iran remained relatively stable from 1990 to 2019. The most recent statistics from 2019 indicate a prevalence rate of around 0.28 in men and 0.25 in women (7, 8).
The protein-protein interaction (PPI) network science field explores different networks and their functions, including biological networks. One example of a biological network is the PPI Network (9). Cellular functions can be categorized into three separate networks: The transcriptional network, the protein interaction network, and the metabolic network. The functionality of a cellular unit is determined by the interconnectedness of these networks, rather than the individual interactions of each network in isolation. This integrated approach is the basis of systems biology, also known as systems biology in Persian.
In biological protein-protein networks, edges represent connections between physically linked proteins, whether directly or indirectly. Various types of biological networks exist, such as the PPI network, which comprises proteins and illustrates their interactions (10). Examination of alterations in gene expression is a common practice in the investigation of diverse human diseases and serves as a method to anticipate intricate cellular and molecular processes linked to such conditions. Given that crucial cellular decisions manifest through shifts in gene expression patterns, the analysis of gene expression proves to be a valuable tool for comprehending the functional components of the genome related to both development and disease (11).
The identification of different genes consistently across studies in diseases such as schizophrenia may represent a meta-analytic starting point for future studies of this type, which researchers can use to examine gene interactions between genes that are altered in expression under the influence of schizophrenia. Analyzing transcripts and the PPI interaction network has the potential to reveal novel pathways relevant to conditions like schizophrenia, and targeting their regulation and inhibition may offer innovative therapeutic strategies (12).
2. Objectives
This study, therefore, scrutinized the protein interaction network associated with schizophrenia through a bioinformatics lens.
3. Methods
This study involved an analytical examination of protein expression data in individuals with schizophrenia as opposed to those in the control group (healthy individuals). Candidate genes included in this study were those previously reported to be associated with schizophrenia based on evidence from at least one of the following study types: In vivo, in vitro, or in silico analyses. Gene and protein data were obtained from publicly accessible databases, including NCBI, GeneCards, SWISS-PROT, and DisGeNET. Additionally, only genes with documented involvement in schizophrenia from prior studies of affected patients and healthy controls were considered.
Genes and proteins lacking prior reported associations with schizophrenia or without supporting evidence from in vivo, in vitro, or in silico studies were excluded from the analysis. Furthermore, no new biological samples were collected, and thus, data not present in existing literature or databases were not included in this study. To achieve this goal, a comprehensive review of literature and exploration of bioinformatics databases such as NCBI, GeneCards, Swiss-Prot, and Diseasome, among others, were conducted to identify genes implicated in the disease using either in vivo, in vitro, or in silico methods. These genes were then selected as candidate proteins, and their expression data were sourced from in vivo, in vitro, and silico studies available in bioinformatics databases.
Following the collection of expression data, the data from each group — cases and controls — were standardized to the control group for comparative analysis and testing of the research hypotheses. Subsequently, the communication network of candidate protein expression data in individuals with the illness and those who are healthy was separately depicted using MATLAB software. The structural parameters of these communication networks from the expression data were then computed and juxtaposed. Identifying statistically significant parameters may unveil promising biomarker candidates.
Through the utilization of text mining techniques, exploring the network interactions among proteins associated with schizophrenia and determining key factors have led to the identification of proteins implicated in the disorder. Subsequently, the array of target proteins associated with this condition was prioritized utilizing the Gene-Disease Association (GDA) score. The expression levels of the categorized candidate genes were retrieved from Gene Entrez and UniProt, and a network depicting the communication among these candidate genes was constructed using the Gephi platform. Furthermore, in addition to delineating the communication network structure among the candidate genes, the network's structural centrality parameters were assessed to pinpoint crucial genes and proteins.
All statistical computations in this research were carried out utilizing R and MATLAB software. For data analysis, sophisticated descriptive and analytical statistical techniques were applied, alongside machine learning methods rooted in advanced bioinformatics algorithms, to compute network data attributes and identify biomarkers associated with network structural characteristics.
4. Results
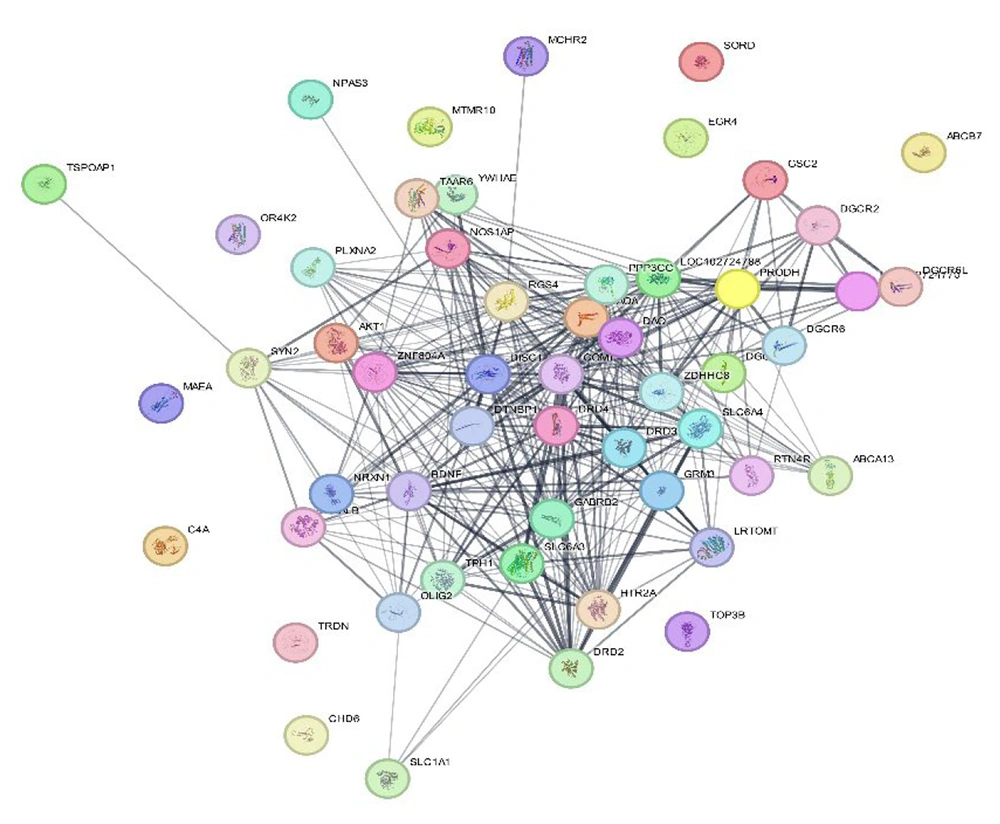
Within this network, the weights of the edges were assigned according to the expression levels of the relevant genes and proteins (Figure 1). Each vertex, such as vertex a, has a specific number of neighbors directly linked to it, denoted as N(a). The MNC score for vertex a is determined by the magnitude of the largest element connected to it. Utilizing this parameter, the top ten biomarkers, as indicated in Table 1, were each assigned the highest respective score (Table 1).
Protein-protein communication network in schizophrenia. Each node represents a protein, and the connecting edge between them is a physical or functional relationship based on at least one type of in vivo, in vitro, or silico study and genes calculated based on the Gene-Disease Association (GDA) criterion.
| Row | Biomarker |
|---|---|
| 1 | COMT |
| 2 | OC102724788 |
| 3 | DISC1 |
| 4 | PRODH |
| 5 | ZDHHC8 |
| 6 | DAO |
| 7 | RGS4 |
| 8 | HTR2A |
| 9 | DAOA |
| 10 | ZNF804A |
MNC Score for Schizophrenia Communication Network
The number of connections linked to a vertex is known as the degree of that vertex. According to this parameter, the top-performing biomarkers within the schizophrenia disease network are outlined in Table 2. Proximity, which assesses a protein's distance to other proteins, is a crucial consideration in identifying biomarkers within biological networks. According to this attribute, the key biomarkers in the schizophrenia disease context are detailed in Table 3.
| Row | Biomarkers |
|---|---|
| 1 | COMT |
| 2 | DISC1 |
| 3 | LOC102724788 |
| 4 | PRODH |
| 5 | ZDHHC8 |
| 6 | RGS4 |
| 7 | DOA |
| 8 | HTR2A |
| 9 | DAOA |
| 10 | ZNF804A |
Degree Score for the Protein Connectivity Network of Schizophrenia
| Row | Biomarkers |
|---|---|
| 1 | DISC1 |
| 2 | LOC102724788 |
| 3 | COMT |
| 4 | PROH |
| 5 | ZDHHC8 |
| 6 | RGS4 |
| 7 | DAO |
| 8 | HTR2A |
| 9 | DAOA |
| 10 | ZNF804A |
Proximity Score for the Protein Connectivity Network of Schizophrenia
The radius, in this context, represents the vertex with the shortest average distance to all other vertices within its immediate network. Based on this "shortest distance" criterion, the highest score was determined for each protein under investigation (Table 4). The betweenness criterion, on the other hand, quantifies the frequency with which a given vertex lies on the shortest paths between other vertices in the network.
| Row | Biomarkers |
|---|---|
| 1 | DISC1 |
| 2 | LOC102724788 |
| 3 | COMT |
| 4 | PRODH |
| 5 | ZDHHC8 |
| 6 | RGS4 |
| 7 | DAO |
| 8 | HTR2A |
| 9 | DAOA |
| 10 | ZNF804A |
Radius Score for the Protein Connectivity Network of Schizophrenia
Due to its position on many shortest paths, a vertex with a high betweenness score is hypothesized to have a significant influence on information flow within the biological network. Consequently, removing such a vertex could disrupt communication throughout the entire network. Applying this principle, the proteins in the schizophrenia disease network were analyzed, and their betweenness scores were calculated (Table 5).
| Row | Biomarkers |
|---|---|
| 1 | DISC1 |
| 2 | COMT |
| 3 | LOC102724788 |
| 4 | RGS4 |
| 5 | ZDHHC8 |
| 6 | SYN2 |
| 7 | PRODH |
| 8 | BDNF |
| 9 | PPP3CC |
| 10 | SLC6A4 |
Bin Score for the Protein Connectivity Network of Schizophrenia
Analysis of the schizophrenia candidate protein communication network, utilizing the top five neighborhood indices (degree, betweenness, proximity, and radius), revealed that disrupted in schizophrenia 1 (DISC1), LOC102724788, catechol-O-methyltransferase (COMT), proline dehydrogenase 1 (PRODH), and zinc finger DHHC-type palmitoyltransferase 8 (ZDHHC8) were consistently identified as key proteins across these measures (Table 6). These proteins exhibited the highest frequency of identification and validation based on these network centrality metrics.
| Mechanism | Diseases Associated with This Protein | Highest Expression Level | Gene Encoding Name | Full Name | Protein Name |
|---|---|---|---|---|---|
| Painkillers, opioids | Childhood-onset schizophrenia | Blood and plasma | DISC1 | Disrupted in schizophrenia 1 | DISC1 |
| Involved in several important enzymatic reactions in metabolism | Junctional brain, Herlitz type | Brain | LOC | MED14-independent group 3 | LOC102724788 |
| Anti-parkinsonian agents, potent COMT inhibitor blocks alpha-synuclein aggregation | Panic disorder syndrome 1 | Nervous system | COMT | Catechol-O-methyltransferase | COMT |
| Redox cofactor (electron carrier), involved in several important enzymatic reactions in metabolism | Deficiency proline oxidase | Bone | PRODH | Proline dehydrogenase 1 | PRODH |
| Anti-parkinson agents | Schizophrenia with or without affective disorder | Plasma | ZDHHC8 | Zinc finger DHHC-type palmitoyltransferase 8 | ZDHHC8 |
Proposed Common Core Proteins Based on 5 Bioinformatic Markers for Schizophrenia
The DISC1 gene, which codes for the DISC1 scaffold protein, exhibits peak expression in blood and plasma and is linked to childhood-onset schizophrenia. Conversely, the LOC102724788 gene, responsible for producing MED14-independent group 3 (LOC), shows its highest expression levels in the brain. The COMT gene, encoding COMT, exhibits the highest expression levels in the nervous system. A link to panic disorder type 1 syndrome has been established, with the proposed mechanism involving the ability of anti-Parkinson's agents, which are potent COMT inhibitors, to prevent the accumulation of alpha-synuclein.
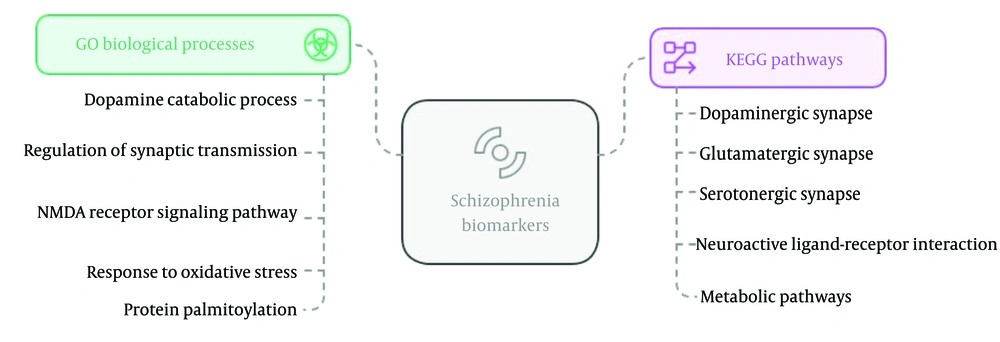
The PRODH gene, which encodes PRODH, is most highly expressed in bone. Proline oxidase deficiency is a disease linked to this gene, arising from a dysfunction in a redox cofactor (electron carrier) crucial for several key enzymatic reactions in metabolism. The ZDHHC8 gene, encoding ZDHHC8, is most highly expressed in plasma. It is associated with schizophrenia, with or without affective disorder, and its functional mechanisms may be relevant to the action of anti-Parkinson agents. Gene ontology (GO) biological process and KEGG pathway enrichment for top-ranked schizophrenia biomarkers are shown in Figure 2.
5. Discussion
Research into protein communication networks in schizophrenia, particularly concerning proteins like DISC1, LOC102724788, COMT, PRODH, and ZDHHC8, aims to elucidate how these proteins interact with each other and intracellular molecules to impact growth and development. Given the strong genetic basis of schizophrenia, a complex neurological disorder, understanding the roles of these proteins in its pathophysiology is critical (13).
Bioinformatics researchers commonly employ various computational tools and databases to investigate the PPI network of schizophrenia-related proteins, such as those discussed previously. The PPI databases, including STRING, IntAct, and BioGRID, facilitate identifying known and predicted interactions between these proteins of interest. To understand the biological context of these proteins and their connection to schizophrenia's development, pathway analysis tools such as KEGG, Reactome, and GO are valuable for identifying the biological pathways in which they are involved (14).
Combining gene expression data from individuals with schizophrenia and control subjects allows for the identification of differentially expressed genes and pathways. Mapping these findings into a protein association network (15) offers valuable insights into the molecular mechanisms driving schizophrenia and may reveal novel targets for therapeutic intervention (16). Computational approaches, including machine learning, offer a powerful means of forecasting novel molecular interactions and pinpointing crucial proteins or pathways implicated in disease mechanisms (17).
Located within a genomically defined active enhancer, validated by STARR-seq (self-transcriptional active regulatory region sequencing), is the protein encoded by LOC102724788 (18). This research also highlights COMT, an enzyme responsible for methylating catecholamines such as the neurotransmitter’s dopamine (DA), epinephrine, and norepinephrine, using S-adenosylmethionine as the methyl donor.
O-methylation represents a primary metabolic route for catecholamine neurotransmitters. Beyond its role in endogenous compound metabolism, COMT plays a critical function in processing catechol-based pharmaceuticals employed in managing conditions like hypertension, asthma, and Parkinson's disease (19). The DISC1 functions as a scaffold protein that modulates neurodevelopmental processes, including neurite outgrowth, synapse formation, and intracellular signaling. The COMT degrades DA and other catecholamines, influencing dopaminergic tone, particularly in the prefrontal cortex, where COMT plays a dominant role due to limited DA transporter expression.
The DISC1 influences DA regulation indirectly through effects on neuronal development and synaptic plasticity, and COMT activity modulates DA levels, which in turn impact DISC1-mediated signaling pathways related to cognition and working memory. Both proteins converge on dopaminergic dysregulation, a key feature in schizophrenia. The DISC1 interacts with DAOA, impacting NMDA receptor function and synaptic integration. Dysfunction in this triad leads to excitatory/inhibitory imbalance, a hallmark in schizophrenia. RGS4 controls signal transduction, with potential regulatory feedback on DISC1-mediated intracellular signaling. This axis supports synaptic maintenance and plasticity, disrupted in schizophrenia.
Madzarac et al. investigated the relationship between COMT (rs4680 and rs4818) and MAO-B (rs1799836 and rs6651806) polymorphisms and the presentation of negative symptoms, including physical and social impairment, in individuals with schizophrenia. Their findings indicated that among female patients, the G allele or GG genotype of COMT rs4680 and rs4818, along with the rs4818-rs4680 GG haplotype (all associated with elevated COMT activity), correlated with greater severity across multiple dimensions of negative symptoms (19).
For male schizophrenia patients, the presence of the MAO-B rs1799836 allele, linked to potentially increased MAO-B activity, correlated with more severe alogia. Additionally, the MAO-B rs6651806 A allele was associated with greater overall negative symptom severity in males. These results suggest a sex-specific relationship between heightened DA degradation, influenced by COMT and MAO-B genetic variants, and the severity of negative symptoms in schizophrenia (19).
Ma et al. also conducted a meta-analysis to test the possible association between the COMT Val158Met polymorphism and antipsychotic response in different populations and antipsychotic types. In this study, there was a highly significant association between COMT Val158Met and antipsychotic response. The accumulated evidence supports the hypothesis that the COMT Val158Met polymorphism affects antipsychotic response in Caucasian and Asian schizophrenia patient populations (20).
The PPI findings reinforce that schizophrenia is a multigenic, multi-pathway disorder. The NMDA receptor hypofunction hypothesis is directly supported by DAO/DAOA findings. DAO inhibitors like Sodium benzoate are already in schizophrenia trials and align well with PPI network insights. Dopaminergic modulation via COMT inhibition (like Tolcapone) offers a strategy to address cognitive deficits, particularly in patients with high COMT activity genotypes. HTR2A antagonism, the core mechanism of atypical antipsychotics, connects directly with network findings, supporting serotonergic dysregulation in schizophrenia.
DISC1, though not directly druggable, modulates multiple interactors in the network. Future small-molecule DISC1 interactome modulators could address neurodevelopmental and synaptic deficits. Multi-target approaches (e.g., combining DAO inhibitors with atypical antipsychotics) might address both positive and cognitive symptoms synergistically.
Ganapathiraju et al. constructed a comprehensive schizophrenia interactome using both experimental (yeast two-hybrid) and literature-curated data. While their network spanned over 3,000 interactions across approximately 190 candidate genes, our analysis focused on high-confidence schizophrenia-associated proteins ranked using multiple centrality metrics, including degree, proximity, radius, and bin scores. Consistently across all metrics, DISC1 emerged as a key hub, reaffirming its critical role in neurodevelopment, synaptic integration, and intracellular signaling. This aligns closely with Ganapathiraju et al.'s identification of DISC1 as one of the most connected nodes in the schizophrenia interactome (12).
Additionally, our network highlighted COMT, DAO, DAOA, and PRODH — proteins less emphasized in earlier studies but with strong therapeutic relevance. For instance, COMT, a catecholamine-degrading enzyme, is modulated by COMT inhibitors like Tolcapone, which have potential implications for cognitive dysfunction in schizophrenia. Similarly, DAO and DAOA are regulators of NMDA receptor function via D-serine metabolism; their inclusion as central proteins supports ongoing trials of DAO inhibitors (e.g., sodium benzoate) as adjunctive treatments (12). These findings align with the current research, underscoring the value of bioinformatics in identifying schizophrenia candidate genes.
The PRODH codes for a protein and is implicated in conditions such as hyperprolinemia, type I, and schizophrenia 4 (SCZD4) (21). The gene in question encodes a mitochondrial protein responsible for initiating proline catabolism. Mutations within this gene are linked to both hyperprolinemia type 1 and susceptibility to SCZD4.
The ZDHHC8, another protein-coding gene, is associated with conditions including prostate cancer and schizophrenia. This gene's GO annotation includes functions related to signal receptors and palmitoyltransferase activity (22). Zhang et al. identified 123 variants in ZDHHC8, comprising five common and 118 rare variants. Of the common variants, rs73198534, rs530313445, and rs74406481 showed significant association with schizophrenia (22). These findings corroborate the results of the current study.
5.1. Conclusions
Based on the results of the analysis of the communication network of candidate proteins in schizophrenia, calculated using the five component indices of the greatest neighborhood, degree, betweenness, proximity, and radius, the proteins DISC1, LOC102724788, COMT, PRODH, and ZDHHC8 had the highest repetition and confirmation rates, respectively. The proteins proposed in this study can be considered as main suggestions for laboratory and clinical study in disease in other neurological and psychiatric disorders.