Introduction
Carotenoids represent a group of valuable molecules because of their valuable pharmaceutical, chemical, coloring and antioxidant properties; several dietary studies have shown that carotenoids combat various types of cancer and other diseases such as cardiovascular disease and cataracts [1, 2]. More than 600 different carotenoids are synthesized by plants and microorganisms; animals cannot synthesize carotenoids, and these pigments must therefore be added to the feeds of farmed species. The synthesis of different natural commercially important carotenoids by several microorganisms such as yeasts of the genera Rhodotorula, Rhordosporidium, Sporobolomyces, Sporidium, Sporobolomyces and Phaffia have led to consider these microorganisms as potential pigment sources [3]. Despite the availability of a variety of natural and synthetic carotenoids there is an interest in microbial sources and yeast as a microorganism with capacity to produce carotenoids, especially Rhodotorula and Phaffia, could be considered as an attractive alternative to the production of carotenoids [3, 4]. Identification and phylogenetic placement of the basidiomycetous yeasts are not always easy, partly because of their polyphyletic nature. The unifying characteristic of these fungi is a predominant unicellular growth phase. Separation of yeasts into the 3 classes of fungi is based on septal morphology, cell wall composition and rDNA analysis. Rhodotorula, an anamorphic genus of heterobasidio- mycetous yeasts is characterized by the following distinctive traits: no ballistoconidia, no fermentation ability, no starch-like compounds and no xylose in whole-cell hydrolyzates [5].
Since the identification of anamorphic (asexual) Basidiomycetous yeasts based on phenotypic characters were considerably difficult, molecular identification is performed. Molecular systematic of yeasts has emphasized either coding (D1/D2 variable domains of the large submit or the complete small subunit (SSU)) or non-coding (maternal transcribed spacers ITS1 and ITS2) regions of the ribosomal DNA. D1/D2 (~600 bases) and the ITS (~600 bases) have received the most attention for yeast systematic [6]. In this present study the strain SG006 was examined by sequence analysis of D1/D2 region, and it investigated the ability of the strain to produce carotenoid. The effect of different medium was considered also.
Materials and Methods
Microorganism Isolation and culture maintenance: In this experimental study the microorganism was isolated on YPG medium (Yeast extract-peptone-Agar (YPG) medium) supplemented with 40 µg/mL of streptomycin and 13 µg/mL penicillin [7]. The origin of the sample which collected from leather waste water was provided by the National Laboratory of Industrial Microbiology, Alzahra University, Tehran, Iran. The red yeast strain was maintained in Petri dish containing YPG agar [yeast extract 5 g/L, agar 17 g/L, peptone 10 g/L, dextrose 20 g/L, and pH=7].
Identification of the microorganism: Three-d-old cultures in YPG agar were used for cellular and colony morphology analysis which carried out by stereomicroscope (LEICA EZ4D, USA). Cell size of 20 randomly selected cells was estimated using microscope (CETi, UK). Conventional tests such as Fermentation, Urea hydrolysis, Diazonium Blue B (DBB), formation of pseudohyphae, starch formation were performed [8, 9].
DNA extraction for PCR was performed following the procedure described by Xiao [10]. NL1 and NL4 were used as universal primers for PCR reaction and the sequencing PCR products [11]. The D1/D2 rDNA gene sequence of the SG006 was compared with those in the National Center for Biotechnology Information (NCBI)/CBS/ nucleotide sequence databases by using the BLAST (blastn) program (http://www.ncbi.nlm.nih.gov), and all of the sequences were aligned using the Clustal W program [12]. A phylogenetic tree and neighbor-joining phylogeny was construct using the MEGA software package, version 5 [13] and bootstrapping was used to estimate the reliability of the phylogenetic reconstructions (1,000 replicates) and only values of 50% or greater were recorded on the resulting tree [14]. The sequence was deposited in the Gene Bank database under the Accession number of JX997835.
Carotenoid analysis: Sample preparation was made by transferring a single colony from the stock culture on YPG agar to 50 mL YPG broth and incubated at 30 overnight. Three mL of the last culture broth (OD 0.50 at 600 nm) was used for inoculation of 100 mL MMS [glucose 10 g/L, (NH4)2SO4 2 g/L, KH2 (PO4) 2 g/L, MgSO4 0.7 g/L, H2O 0.5 g/L, CaCl2 0.2 g/L, H2O 0.1 g/L, yeast extract 1 g/L, pH=5] in 500 mL Erlenmeyer flask and aerated in a shaker incubator at 150 rpm and 30ºC for 72 h. All the experiments were carried out independently in triplicates and the results presented here are the mean of the three. After cultivation the cells were harvested by centrifugation at 10.000 rpm for 20 min and washed three-time with distilled water and centrifuged again. The obtained biomass at first was held in -70ºС for 24 h and then was transferred to 35ºС for 24 h. The methods of Davis [15] with modification were used for the extraction of carotenoid pigments. Briefly, cells were harvested by centrifugation at 10,000 rpm for 10 min and were washed 3 times with distilled water. Cells were ruptured 3 times with 12 mL of acetone and broken using homogenizer (Witeg, Germany).
The suspension was then centrifuged and the supernatant collected. Acetone extracts were pooled in a funnel and carotenoid pigments were extracted twice with an equal volume of petroleum ether. For analytical method pigments were measured by using spectrophotometer at 450 nm using the extinction coefficient E1%450=2500. Cell dry weight was determined after heating them at 105ºC to a constant weight. The ability to produce biomass and carotenoid pigments by the strain was further assessed using 2 alternative culture MMS and YM [yeast extract 3 g/L, malt extract 3 g/L, peptone 5 g/L, and glucose 10 g/L, pH=5], at the same condition as described above.
Statistical analysis: All statistical analyses were performed with using SPSS-18 programs. Data from the experiments were subjected to student t-test. Values p<0.001 were considered to be significant.
Results
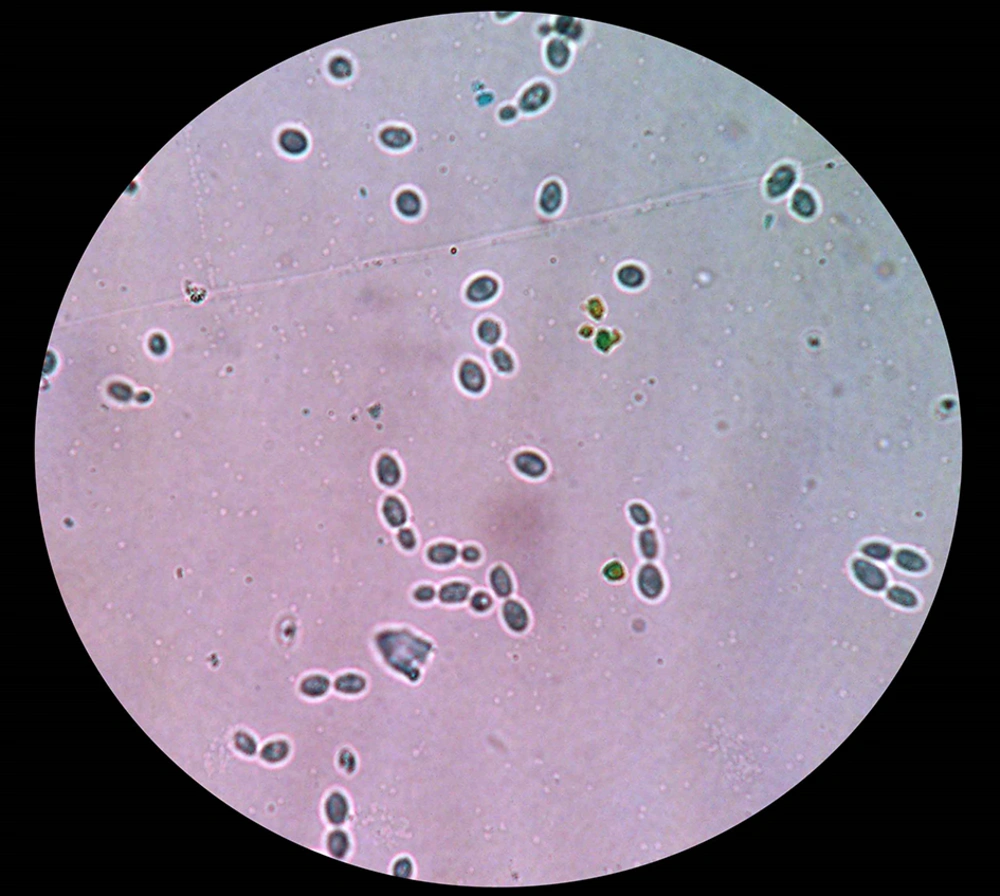
The local isolate of the yeast-like microorganism, Rhodotorula slooffiae strain SG006, was isolated from a sample which collected from leather wastewater in Iran. The isolate developed mucous, smooth surface and light salmon-colored colonies on YPG medium and budding was frequently observed in microscopic examination (Fig. 1, 2). The size of spherical cells of SG006 strain was 6.83±0.14 μm and colony diameter was 1.5 mm. The positive result of DBB test indicated that SG006 belongs to Basidiomycetous yeast. Starch-like substances was not synthesized by the strain. Sugars such as glucose, fructose, mannose, sucrose, lactose and galactose were not assimilated by SG006 strain, i.e., the ability of fermentation was lacked. Pseudohyphae were not developed in this strain but urease activity was found.
Molecular characterization: After using the Blast program, R. slooffiae strain 2 (AB566328.1) with 100% identities, R. slooffiae strain 9 (EU583485.1) with 100% identities and R. slooffiae isolate 1-2-8 (AF514865.1) with 99% identities had the closest similarity to tested strain. A phylogenetic tree which drew from neighbor-joining analysis based on sequences of the 26S rDNA D1/D2 domain has been shown in figure 3. The sequence was deposited in the Gene Bank database under the Accession number of JX997835.
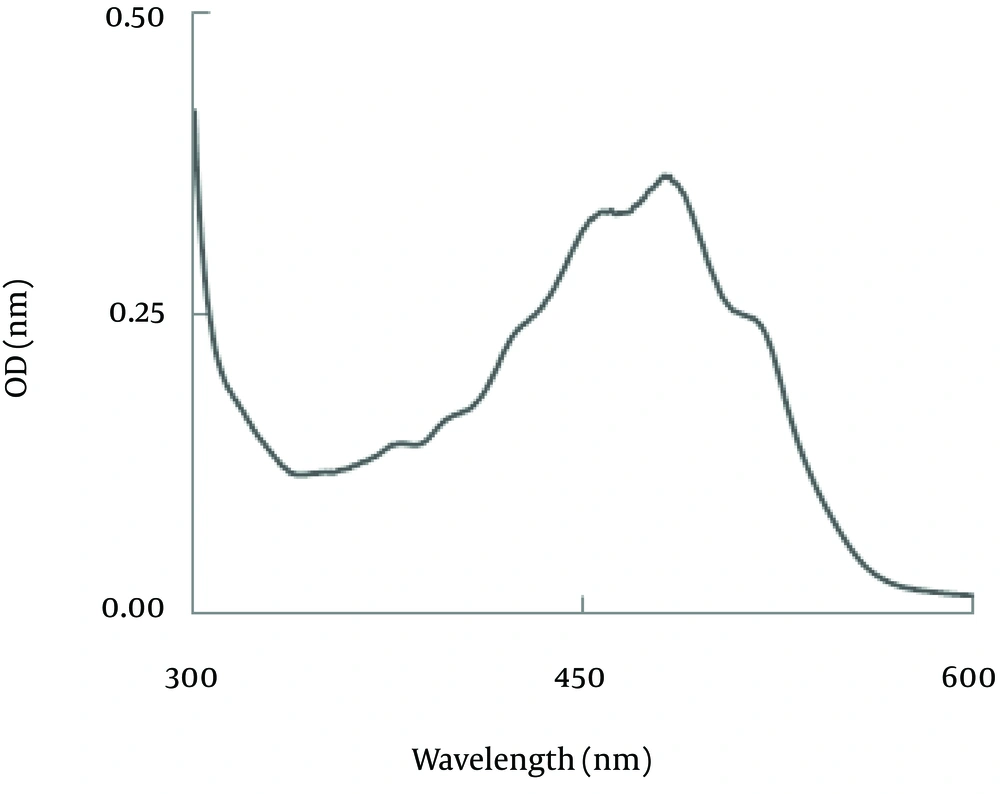
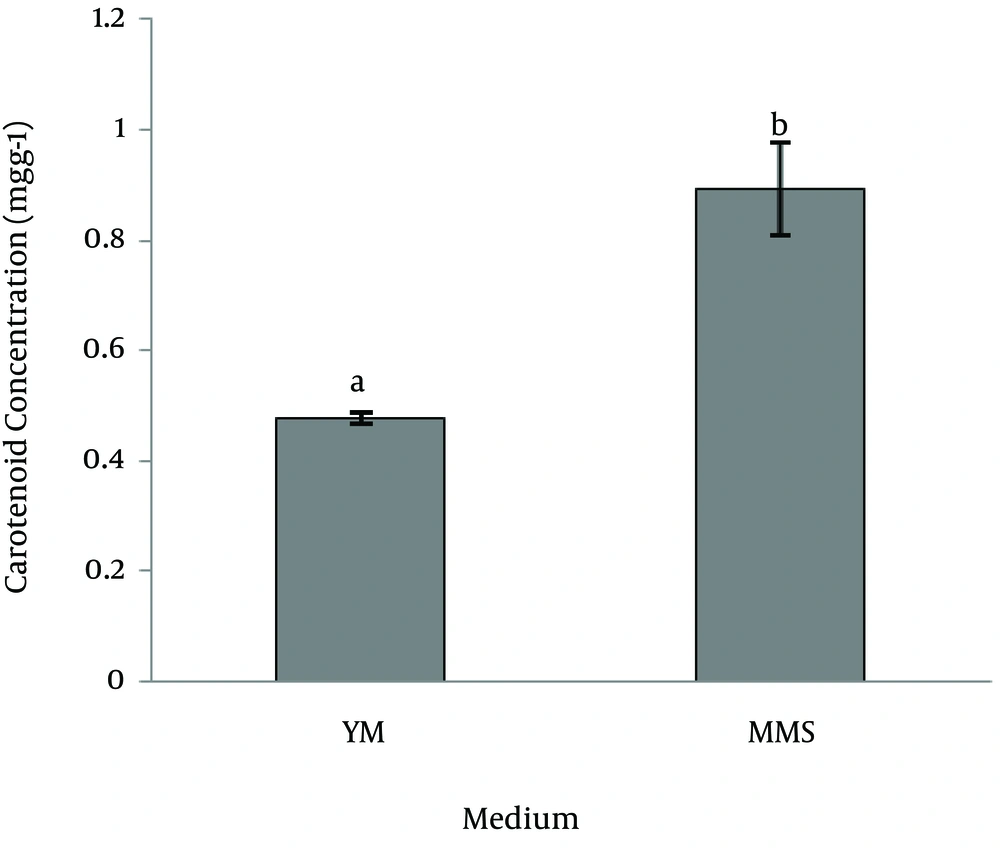
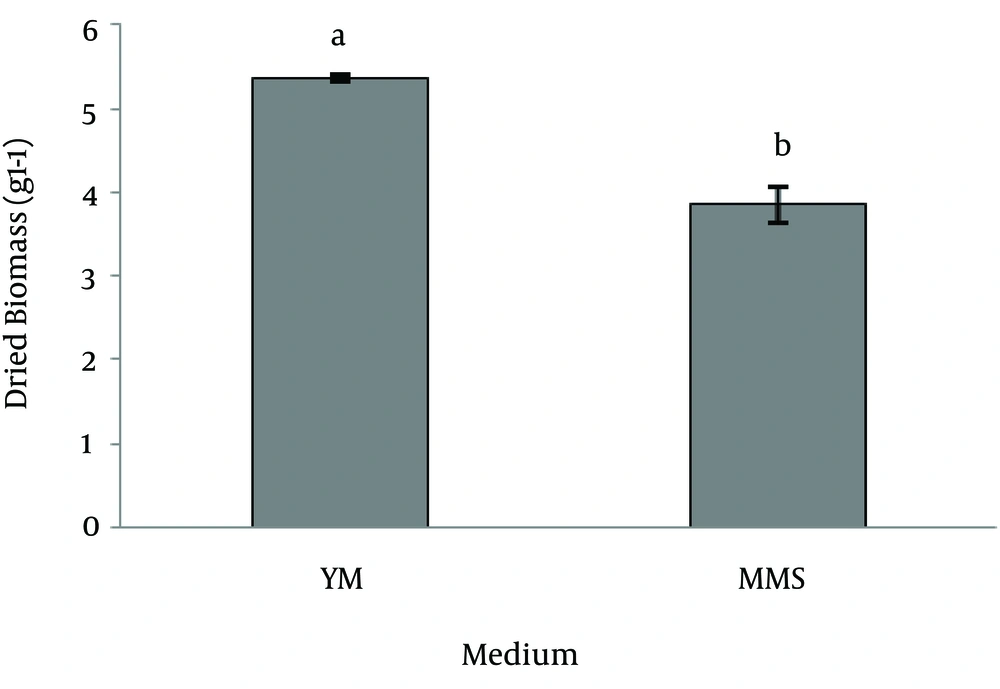
Carotenoid analysis:Figure 4 showed the spectrum of an extract obtained from a strain SG006, with maximum absorption at 457, 482, 516 nM in petroleum ether. The result of Pigments measurement which evaluated spectro- photometrically at 450 nm using the extinction coefficient E1%450=2500 showed that the amount of carotenoid concentration was 0.084±4.672 (mg/g) and dry cell was 0.367±3.5 (g/L). Production of carotenoid in MMS medium was significantly higher than YM medium whereas the biomass accumulation in YM medium was significantly higher (p<0.001). The results have been illustrated in figures 5, 6.
Discussion
The present study indentified the strain SG006 as R. slooffiae based on the sequence analysis of the D1/D2 region in addition to morphological and biochemical characterization; besides, it demonstrated that the strain was capable to produce carotenoid.
The approach to yeast identification has significantly changed in just a few decades due to rapid increase in basic biological knowledge, increased interest in the practical applications and biodiversity of this important microbial group, and enormous technological advances especially in the sphere of molecular tools. While some conventional methods are still tenable, many molecular techniques have been developed that allow for strain classification at all taxonomic levels. However, the oldest tool of microbiology, the microscope, is still a fundamental accessory for studies involving yeast biology, biodiversity and taxonomy [16]. Lack of fermentation ability, formation of starch-like compound and formation of hyphae or Pseudohyphae, as well as presence of urease activity are important criteria in the taxonomy and identification of Rhodotorula sp; as described by Barnett et al. [9]. Our biochemical results placed SG006 in Basidiomycetous yeasts which are concordant with the standard description of Rhodotorula sp. In addition, molecular identification by the 26S rDNA D1/D2 domain confirmed our finding. The pigmented yeasts of the genera Rhodotorula, Rhordosporidium, Sporobolomyces have the capacity to produce carotenoid [17]. One of the most important characteristics of a carotenoid is its electronic absorption spectrum [18]. This is a function of the chromophor, so the presence of the three-peak spectrum in visible absorption spectra of the sample determined it as carotenoids. Several studies examined the carotenoid production of Rhodotorula species. Carotenoid production in present study (4672±0.084 μg/g) was higher than that reported for Rhodotrula spp. by other authors under different culture condition [19- 22] Martin et al. [23] reported 1.256 μg/g for the total carotenoid concentration of R. slooffiae which is lower than our concentration (4672 μg/g). Meanwhile, Frengova et al. reported 14.3 g/L and 2.67 mg/L for dried biomass and total carotenoid production, respectively [24]
Such data discrepancy between different studies may be explained by differences in medium composition and experimental conditions. R. slooffiae has higher potential to carotenoid production than R. Mucilaginosa under the same condition; although it was demonstrated that the highest value of carotenoids are produced after 48 h incubation (unpublished data). We found that MMS medium promoted carotenoid production rather than biomass accumulation whereas the highest amount of biomass was obtained by growth in YM broth. Our results are confirmed by data of Buzzini et al. [25], Fang and Chiou [26], Johnson and An [27]. They have shown that the maximum values of total carotenoids are not directly correlated to the maximum value of cell biomass. Asku and Tugba-Eren [28] and Voaides et al. [29] have pointed out that the types of carotenoids and their relative amount may vary depend on the cultivation medium. They determined that the carotenoid contents in the cell mass reached their maximums when the cell growth in MMS medium rather than YM, a fact that can be confirmed also by the data obtained from this study.
Sequence analysis of the D1/D2 region in addition to morphological and biochemical characterization represented strain SG006 as a R. slooffiae and it indicated that SG006 is capable to produce carotenoids, although the internal transcribed spacer region is required to distinguish closely related species. The intergenic spacer region is recommended for additional differentiation of species and strains. The results from this study would be of significance for the carotenoid industries to introduce potential microorganism to produce carotenoid pigments naturally. Nevertheless, the carotenoid production by this strain in large scale will be required more detailed studies.