1. Introduction
Maple syrup urine disease (MSUD; OMIM ID # 248600) is an autosomal recessively inherited error of metabolism resulting from the deficiency in the mitochondrial branched-chain alpha keto acid dehydrogenase complex caused by mutations in one of the four genes BCKDHA, BCKDHB, DBT and DLD. There are at least five main clinical phenotypes in MSUD patients differing in presentation and severity of the disease. The majority of affected individuals (~75%) have the classic form of the disease in which the residual enzymatic activity is near 2% of normal activity [1-6].
The estimated worldwide incidence of MSUD ranges from 1 in 185,000 to 1 in 940,000 in western countries; the prevalence is much higher in certain ethnic groups, specifically the Mennonite populations of Pennsylvania and other nearby states (1 in 176) [7].
We report here a novel homozygote missense mutation in the BCKDHB gene in the province Khuzestan.
2. Methods
2.1. Patient
The patient was a three years old girl, one of two children of consanguineous parent. The patient exhibited the classical clinical phenotype of MSUD. The medical diagnosis could only be completed later by the plasma amino acid analysis high BCAAs have been observed, with leucine at 1970 μmol/L (normal range 55 - 167 μmol/L), isoleucine at 1978 μmol/L (normal range 52 - 283 μmol/L). After obtaining an informed consent, all family members were questioned about their personal medical history.
2.2. Sample Collection, DNA Isolation and Polymerase Chain Reaction
Peripheral blood was collected from patient and her parents in the EDTA tubes. The DNA was extracted by the standard salting out protocol, and the quality of the extracted DNA was verified by gel electrophoresis. All coding exons of the BCKDHA, BCKDHB, DBT and DLD genes were amplified by PCR using primers designed by software Primer3 (http://primer3.ut.ee/). Primers are showed in Table 1.
| Exon | Forward Primer | Reverse Primer | Amplicon Length, bp | Tm, °C |
|---|---|---|---|---|
| 1 | 5'-GCCACCTCTAGCCCACACTTCC-3' | 5'-AATAAGCTGGGATGCAAGGA-3' | 438 | 59.6 |
| 2 | 5'-TGTATTTTACAACACACGAAATGA-3' | 5'-TGTTAACCATTGAGCTCCACA-3' | 216 | 59.0 |
| 3 | 5'TGCACTGTTGGCTTGCGAGACAA-3' | 5'-TTCTGCGGGTGGCGTTGGAA-3' | 170 | 60.1 |
| 4 | 5'-CCTTGATCGAGATCTATGGTCC-3' | 5'-TCCATACAATGGGGATATGACA-3' | 307 | 60.3 |
| 5 | 5'-TACCACCATCCATCACCAGA-3' | 5'-TGAATAAGGCAGTCACAAAAGG-3' | 192 | 61.2 |
| 6 | 5'-CATGACATTACTCTCATTTGCCA-3' | 5'-GGGTAGCGGCAATACTTGAA-3' | 250 | 59.6 |
| 7 | 5'-TGCACAAGTGTCACCTCAGA-3' | 5'-GCTTCCAAGCACAACGTAGG-3' | 205 | 59.7 |
| 8 | 5'-CCACATGAATTGATTGTGAGC-3' | 5'-CAAAAATAACTTGCCTCGCA-3' | 176 | 59.4 |
| 9 | 5'-ACCTGTCGAAAGCGAGTTGT-3' | 5'-GGAATTGCACAAGCATTGAA-3' | 294 | 59.7 |
| 10 | 5'-GGATCATGCGAACATGCTGTTACC-3' | 5'-ACACTGATGATTGCTGTGTCTTGG-3' | 402 | 60.3 |
| 11 | 5'-GCATTCAACTAGTTTTTGAGGC-3' | 5'-GCCAAAGGTTTCAGGGAAAT-3' | 226 | 59.5 |
Sequences of the Forward and Reverse Primers Used for Separate Amplification of Exons in the BCKDHB Gene
The PCR was performed in the following conditions: 100 ng genomic DNA, 200 μM dNTP, 1.5 mM MgCl2, 2.5 units Super Taq polymerase, and 25 pmoL of each primer. Amplification was carried out in a total volume of 25 μL. After an initial denaturation at 93°C for 5 minutes, followed by 35 cycles of 93°C for 1 minute, annealing at 60°C for 30 seconds, and extension at 72°C for 45 seconds, and ended by a final extension at 72°C for 3 minutes.
PCR products were directly sequenced and analyzed by the ABI Prism 3700 automated genetic analyzer (Applied Biosystems). The results were analyzed and provide with Chromas, Fast-PCR, and compared with the reported gene sequence using the BLASTN program.
3. Results
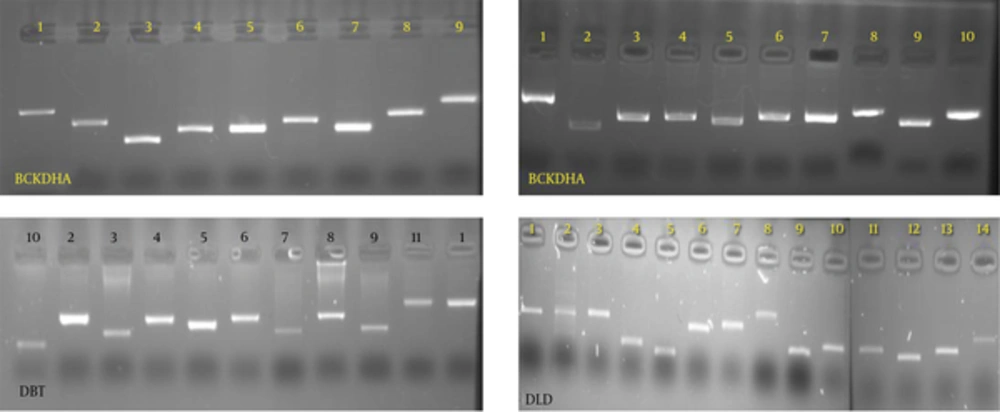
Exons and the flanking intron sequences were amplified for each four genes. Figure 1 demonstrated PCR products for exons of the subjected genes.
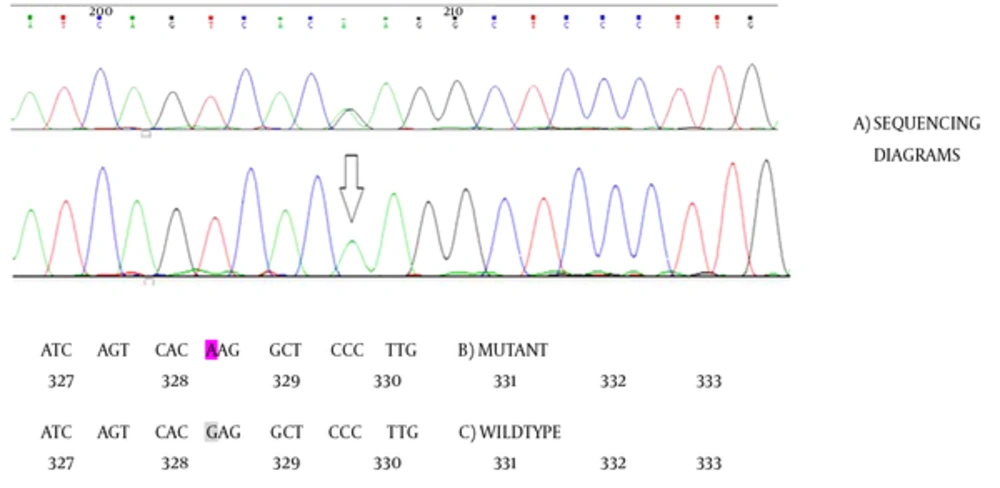
Direct sequencing analysis of the patient and her parents after comparison with the NCBI reference sequence of the BCKDHA, the BCKDHB, the DBT, and the DLD genes demonstrated a homozygous missense mutation GAG → AAG substitution in the BCKDHB gene at exon 9, which causes an amino acid exchange of glutamate to lysine at codon 330 (Figure 2). This mutation has not been previously reported. Samples from parent showed the mentioned change in a heterozygous manner. We further evaluate the pathogenic properties of the mentioned change by different mutation prediction programs, which predicted it as pathogenic. DNA samples from 50 individuals were tested in order to confirm the absence of new identified mutation. The prediction of the functional effect of the missense single nucleotide alteration was done using software PolyPhen2 (Polymorphism Phenotyping v2) [8], SIFT (Sorting Intolerant From Tolerant) [9], Mutation taster [10], and Predict SNP [11, 12]. Results are summarized in the Table 2.
| Program | Result | Score of Detected Change in This Report | Benign Score | Damaging Score |
|---|---|---|---|---|
| Sift | Damaging | 0.0 | 1.0 | 0.0 |
| Polyphen2 | Damaging | 1.0 | 0.0 | 1.0 |
| Mutation Taster | Pathogen | > 0.5 | < 0.5 | Close to 1 |
| Snap | Non-Natural | 85% | < 50% | 100% |
| Mutation Assessor | pathogen | 92% | < 50% | 100% |
Diverse Prediction Programs Have Been Used for Improving Pathogenic Nature of the Novel Change in the BCKDHB Gene. The Last Two Columns Show the Scoring of Benign Versus Damaging Prediction. According Mentioned Prediction Programs, the Novel Mutation is Pathogen With High Probability
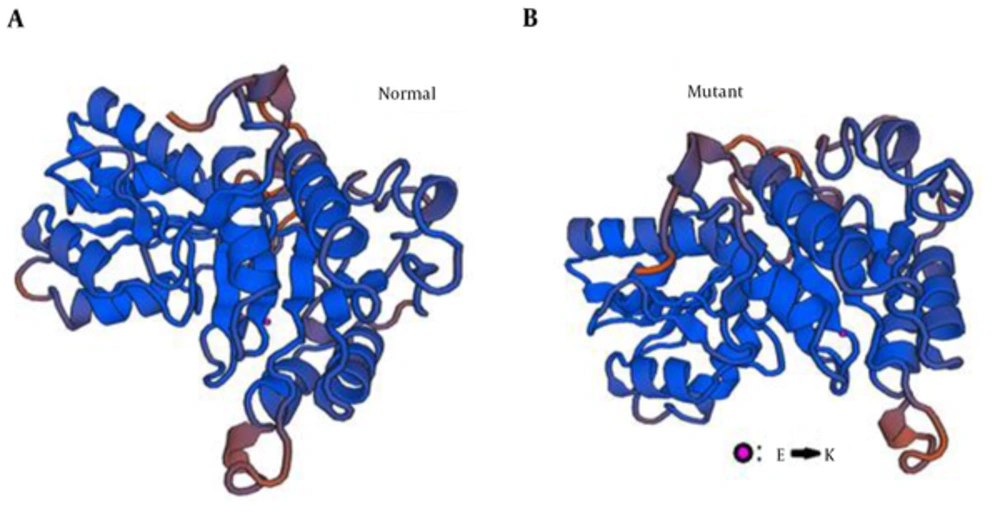
In addition, bioinformatics analysis by Swiss modeling software (www.swiss-prot.org) reveals that the mutant amino acid causes conformational changes in the sequence of BCKDHB gene. As visible in Figure 3, it changes the β-sheet conformation at first and totally effects whole protein structure.
4. Discussion
As described, a homozygous variant was identified in the BCKDHB gene in the affected girl. It was a missense mutation G > A (E330k-β) located in exon 9 (c.1110G > A) leading to an amino acid substitution of glutamate for lysine at position 330. Her parents were heterozygous for this mutation. Afterwards, for verifying the Pathogenicity of this mutation, PCR and direct sequencing was performed for exon 9 and its flanking regions in 50 unrelated individuals. Neither mutation was found in the 100 alleles of 50 normal individuals that were in the same ethnicity as the patient’s family from Southwest Iran.
To date and regarding Human Genome Database (www.HGMD.org) , more than 187 mutations have been found in the four MSUD causing genes BCKDHA, BCKDHB, DBT, and recently identified DTT gene responsible for mild form of MSUD. But, most reported mutations occur in the BCKDHA or the BCKDHB gene with 45 and 35 percents in MSUD patients worldwide, respectively (Genetics Home Reference, ww.ncbi.nlm.nih.gov).
According recent report, it seems that occurrence of some mutations in the MSUD causing genes depends on ethnicities, so that targeted mutations could be suggested for individuals of certain geographic locations [13].
Divers mutation prediction programs suggested the novel change as pathogen (Table 2) with max score indexing. That means the highest probability for Pathogenicity. Furthermore, 3D structure protein model was created. Hereby, mutant versus wild type model was compared. As in the figure 2 has been showed, the conformation change might negatively affects the complex building between the BCKDH and its partners.
Considering the above mentioned findings, and clinical evidence of MSUD diagnosis in the patient we conclude that the nucleotide change is causative for the outbreak of disease and will extend the mutation list of MDUD patients. This is significant for screening of affected individuals at least in southwest Iran. Despite, functional study could confirm definitely the Pathogenicity of the detected change in our patient.
Finally, identification of the novel mutation shed some light on the molecular pathology of MSUD in Iranian subjects. In the meantime, the detection of the novel mutation in the Iranian patient would facilitate prenatal identification in her at-risk family.