1. Background
Klebsiella pneumoniae is a member of the Enterobacteriaceae family. K. pneumoniae strains are ubiquitous in nature and can be found in natural environments and on mucosal surfaces of mammals [1, 2]. This microorganism is an opportunistic bacterial pathogen that plays a main role in most human infections such as urinary tract infections, respiratory tract infections, gastrointestinal tract infections etc. [1].
Extended-spectrum beta-lactamases (ESBLs) are bacterial enzymes that confer resistance to penicillins, broad-spectrum oxyimino-cephalosporins and aztreonam, but inhibited by β-lactamase inhibitors such as clavulanic acid [3].
K. pneumoniae is one of the major ESBLs-producing bacteria [4]. Today, ESBLs-producing K. pneumoniae is listed as one of the six drug-resistant pathogens that can be caused dangerous infections in humans for which few potentially effective drugs are available [5].
Among ESBLs-producing strains, co-resistance with other classes of antibiotics such as aminoglycosides, fluoroquinolones, tetracyclines and sulfonamides are also widespread [6].
2. Objectives
The aim of this study was to determine the antibiotic resistance pattern and co-resistance of ESBLs-producing K. pneumoniae clinical isolates at three major hospitals of Zahedan by phenotypic methods.
3. Methods
In this sectional-descriptive study, fifty one ESBLs-producing isolates of K. pneumoniae were obtained from a collection previously described [7]. Bacteria were isolated from patients at three major hospitals (Ali ibn Abi Talib, Khatam al Anbiya and Buali) of Zahedan during November 2011 to November 2012. These isolates were recovered from urine (n = 37), respiratory tract secretions (n = 9), blood (n = 1), urinary bladder catheter (n = 2), tracheal fluid (n = 1) and joint fluid (n = 1).
ESBLs production was confirmed using the phenotypic confirmatory test. Susceptibility of the isolates to 18 antimicrobial agents were tested by Kirby-Bauer disk diffusion assay and the results were interpreted according to the CLSI criteria [8]. The used antibiotic disks (MAST, Merseyside, UK) were included cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cefpodoxime (10 μg), cefepime (30 μg), aztreonam (30 μg), imipenem (10 μg), gentamicin (10 μg), streptomycin (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), nalidixic acid (30 μg), nitrofurantoin (300 μg), tetracycline (30 μg), chloramphenicol (30 μg), colistin sulfate (25 μg), amoxicillin (25 μg) and trimethoprim/sulfamethoxazole (1.25/23.75 μg). Escherichia coli ATCC 25922 was used as quality control for antibiotic disks.
Minimum inhibitory concentration (MIC)-values of four antibiotics consist of cefotaxime, ceftazidime, ceftriaxone and cefpodoxime were determined for isolates by the E-Test method (Liofilchem MIC test strips, Italy). Concentration gradient for cefotaxime was 0.002 - 32 µg/mL, and for ceftazidime, ceftriaxone and cefpodoxime were 0.016 - 256 µg/mL. Interpretive criteria used were as per the E-test manufacturer’s guidelines and CLSI. E. coli ATCC 25922 was used as quality control for MIC test strips [9].
All descriptive statistical analyses were performed by applying the SPSS software for Windows, version 18.0 and Excel 2013 software.
4. Results
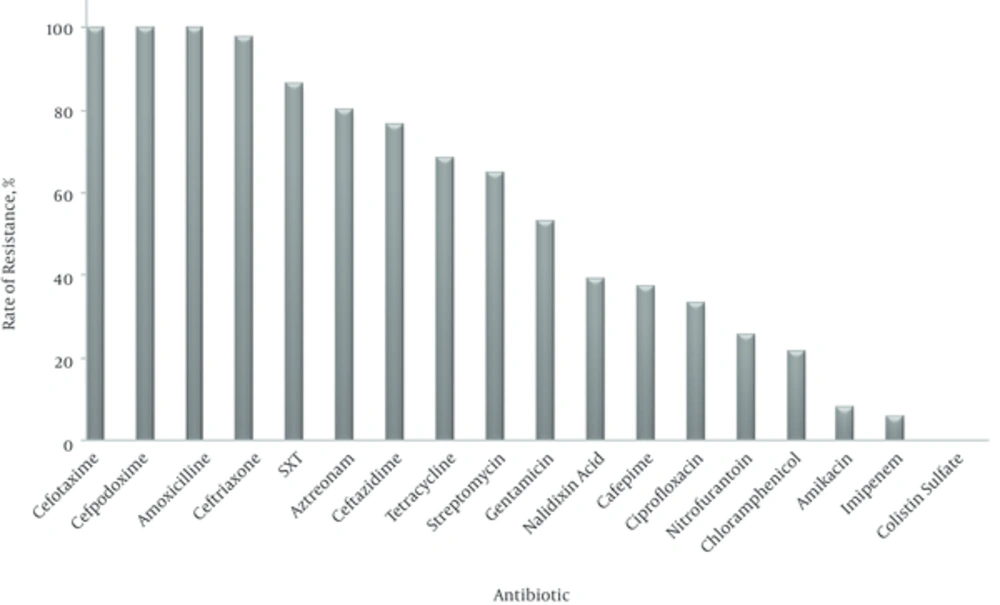
The antibiotic resistance pattern of isolates is shown in Figure 1. As observed, all isolates were resistant to cefotaxime, cefpodoxime and amoxicillin followed by 98% resistance to ceftriaxone, 86.3% to trimethoprim/sulfamethoxazole, 80.4% to aztreonam, 76.5% to ceftazidime, 68.6% to tetracycline, 64.7% to streptomycin, 52.9% to gentamicin, 39.2% to nalidixic acid, 37.3% to cefepime, 33.3% to ciprofloxacin, 25.5% to nitrofurantoin, 21.6% to chloramphenicol, 7.8% to amikacin and 5.9% to imipenem. No isolates were resistant to colistin sulfate. The most active antibiotic was colistin sulfate followed by imipenem and amikacin.
The disk diffusion assay revealed that all isolates had co-resistance to several antibiotics. Characteristics of these isolates and their MIC-values were demonstrated in Table 1.
| Isolate Number | Specimen | Resistance and co-Resistance of ESBLs-Producing Isolates | MIC, μg/mL | |||
|---|---|---|---|---|---|---|
| Cefotaxime | Ceftazidime | Ceftriaxone | Cefpodoxime | |||
| 2 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-C-GM-A | 32 < | 24 | 256 < | 256 < |
| 6 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-C-GM-A | 32 < | 24 | 256 < | 256 < |
| 12 k | Respiratory tract secretions | CTX-CRO-CPD-GM-SXT-TE-A | 32 < | 3 | 48 | 256 < |
| 13 k | Urine | CTX-CAZ-CRO-CPD-ATM-SXT-A | 32 < | 8 | 96 | 256 < |
| 16 k | Urine | CTX-CAZ-CRO-CPD-ATM-SXT-TE-A | 32 < | 16 | 6 | 256 < |
| 22 k | Urine | CTX-CAZ-CRO-CPD-ATM-C-SXT-TE-A | 32 < | 256 < | 96 | 256 < |
| 36 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-AK-NA-S-SXT-TE-A | 32 < | 16 | 256 < | 256 < |
| 45 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-FM-A | 32 < | 24 | 256 < | 256 < |
| 46 k | Respiratory tract secretions | CTX-CAZ-CRO-CPD-ATM-CPM-NA-CIP-GM-S-SXT-TE-A | 32 < | 48 | 256 < | 256 < |
| 48 k | Tracheal fluid | CTX-CAZ-CRO-CPD-ATM-AK-NA-SXT-TE-A | 32 < | 16 | 256 < | 256 < |
| 49 k | Urine | CTX-CAZ-CRO-CPD-ATM-NA-GM-S-SXT-TE-A | 32 < | 12 | 64 | 256 < |
| 53 k | Respiratory tract secretions | CTX-CAZ-CRO-CPD-ATM-CPM-IMI-NA-CIP-GM-S-FM-SXT-TE-A | 32 < | 256 < | 256 < | 256 < |
| 56 k | Urine | CTX-CAZ-CRO-CPD-ATM-SXT-TE-A | 32 < | 8 | 48 | 256 < |
| 61 k | Urine | CTX-CAZ-CRO-CPD-ATM-SXT-TE-A | 32 < | 8 | 64 | 256 < |
| 63 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-C-AK-NA-S-SXT-TE-A | 32 < | 8 | 192 | 256 < |
| 67 k | Respiratory tract secretions | CTX-CAZ-CRO-CPD-ATM-CPM-IMI-NA-CIP-GM-S-FM-SXT-TE-A | 32 < | 256 < | 256 < | 256 < |
| 68 k | Urinary bladder catheter | CTX-CAZ-CRO-CPD-ATM-CPM-IMI-NA-CIP-GM-S-FM-SXT-TE-A | 32 < | 48 | 256 < | 256 < |
| 71 k | Respiratory tract secretions | CTX-CRO-CPD-ATM-NA-CIP-GM-S-FM-SXT-TE-A | 32 < | 4 | 48 | 256 < |
| 74 k | Urine | CTX-CAZ-CRO-CPD-ATM-S-SXT-TE-A | 32 < | 8 | 64 | 256 < |
| 77 k | Urine | CTX-CAZ-CPD-ATM-A | 2 | 12 | 2 | 256 < |
| 78 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-C-NA-CIP-GM-S-SXT-TE-A | 32 < | 48 | 256 < | 256 < |
| 93 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-NA-CIP-GM-S-FM-SXT-TE-A | 32 < | 24 | 256 < | 256 < |
| 97 k | Urine | CTX-CAZ-CRO-CPD-ATM-NA-CIP-GM-S-SXT-TE-A | 32 < | 16 | 64 | 256 < |
| 114 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-S-FM-SXT-A | 32 < | 48 | 256 < | 256 < |
| 119 k | Joint fluid | CTX-CAZ-CRO-CPD-ATM-GM-S-SXT-A | 32 < | 8 | 96 | 256 < |
| 120 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-SXT-A | 32 < | 12 | 256 < | 256 < |
| 130 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-C-NA-S-SXT-TE-A | 32 < | 32 | 256 < | 256 < |
| 131 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-CIP-GM-S-SXT-TE-A | 32 < | 256 < | 96 | 256 < |
| 136 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-GM-SXT-A | 32 < | 96 | 256 < | 256 < |
| 143 k | Respiratory tract secretions | CTX-CAZ-CRO-CPD-ATM-CPM-GM-S-SXT-TE-A | 32 < | 96 | 256 < | 256 < |
| 146 k | Urine | CTX-CAZ-CRO-CPD-ATM-S-SXT-A | 32 < | 12 | 96 | 256 < |
| 147 k | Urine | CTX-CAZ-CRO-CPD-ATM-C-CIP-GM-S-SXT-TE-A | 16 | 4 | 48 | 256 < |
| 148 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-NA-GM-S-SXT-A | 32 < | 16 | 96 | 256 < |
| 150 k | Urine | CTX-CAZ-CRO-CPD-ATM-C-NA-S-SXT-TE-A | 32 < | 6 | 64 | 256 < |
| 162 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-IMI-NA-CIP-GM-S-FM-SXT-TE-A | 32 < | 256 < | 256 < | 256 < |
| 165 k | Urine | CTX-CRO-CPD-ATM-C-NA-CIP-GM-S-FM-A | 32 < | 6 | 96 | 256 < |
| 169 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-NA-S-SXT-TE-A | 32 < | 12 | 96 | 256 < |
| 191 k | Respiratory tract secretions | CTX-CRO-CPD-NA-CIP-GM-SXT-TE-A | 32 < | 6 | 128 | 256 < |
| 194 k | Urine | CTX-CAZ-CRO-CPD-ATM-NA-S-SXT-TE-A | 32 < | 8 | 96 | 256 < |
| 195 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-CIP-GM-S-SXT-TE-A | 32 < | 16 | 256 < | 256 < |
| 196 k | Respiratory tract secretions | CTX-CRO-CPD-C-NA-CIP-GM-S-FM-SXT-TE-A | 32 < | 6 | 128 | 256 < |
| 198 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-C-NA-CIP-GM-FM-SXT-TE-A | 32 < | 96 | 256 < | 256 < |
| 199 k | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-NA-CIP-FM-A | 32 < | 48 | 256 < | 256 < |
| 3 a | Urine | CTX-CRO-CPD-S-SXT-TE-A | 32 < | 6 | 64 | 256 < |
| 17 a | Urine | CTX-CRO-CPD-ATM-S-SXT-TE-A | 32 < | 3 | 64 | 256 < |
| 58 a | Urine | CTX-CAZ-CRO-CPD-ATM-CPM-NA-CIP-GM-S-SXT-A | 32 < | 16 | 256 < | 256 < |
| 106 b | Urine | CTX-CAZ-CRO-CPD-ATM-NA-SXT-TE-A | 32 < | 12 | 96 | 256 < |
| 107 b | Respiratory tract secretions | CTX-CAZ-CRO-CPD-ATM-CPM-AK-GM-S-A | 32 < | 24 | 256 < | 256 < |
| 126 b | Urinary bladder catheter | CTX-CAZ-CRO-CPD-ATM-NA-CIP-GM-S-SXT-TE-A | 32 < | 8 | 64 | 256 < |
| 130 b | Urine | CTX-CRO-CPD-S-SXT-TE-A | 32 < | 6 | 48 | 256 < |
| 133 b | Blood | CTX-CAZ-CRO-CPD-ATM-CPM-S-FM-SXT-A | 32 < | 12 | 256 < | 256 < |
Abbreviations: a, Ali ibn Abi Talib hospital; AK, Amikacin; A, Amoxicillin; ATM, Aztreonam; b, Buali hospital; CTX, Cefotaxime; CAZ, Ceftazidime; CRO, Ceftriaxone; CPD, Cefpodoxime; CPM, Cefepime; C, Chloramphenicol; CIP, Ciprofloxacin; FM, Nitrofurantoin; GM, Gentamicin; k, Khatam al Anbiya hospital; IMI, Imipenem; NA, Nalidixic acid; S, Streptomycin, SXT; Trimethoprim/sulfamethoxazole; TE, Tetracycline.
5. Discussion
In this study, all ESBLs-producing isolates were resistant to amoxicillin and susceptible to colistin sulfate. Moreover, most isolates were resistant to trimethoprim/sulfamethoxazole and aztreonam. As ESBLs-encoding genes are generally found on multi-drug resistance plasmids, many of ESBLs-producing isolates also are resistant to other classes of antibiotics such as tetracyclines, aminoglicosides, flouroquinolons, sulfonamides and chloramphenicol [6, 10]. In our study, we obtained the similar results: the rate of resistance against tetracycline, aminoglicosides and flouroquinolons was higher compared to non-ESBLs-producing isolates in similar studies [11, 12].
Saeidi et al. [13], in their study on ESBLs-producing K. pneumoniae isolates in Zabol (Southeast Iran) have shown that the highest antibiotic resistance was against beta-lactams, sulfonamides and aminoglicosides which in parallel with our results.
Among the third-generation cephalosporins, the highest resistance was observed against cefotaxime and cefpodoxime with 100% and ceftriaxone (98%); and the lowest resistance was observed against ceftazidime (76.5%). Our findings suggest among the beta-lactam antibiotics, carbapenems (e.g. imipenem) are the most effective drugs for ESBLs-producing K. pneumoniae strains. Several investigations support our findings [12, 14].
In this study, the high sensitivity of isolates to colistin sulfate, imipenem and amikacin may be due to limited use of these antibiotics in patients.
As demonstrated in Table 1, of 18 antibiotics tested, the ESBLs-producing isolates were resistant to the minimum and maximum number of 7 and 15 antibiotics, respectively. These results proposes the transfer of multi-drug resistance plasmids among Gram-negative bacteria [15], thus endangering the treatment of infections caused by these strains.
In summary, the results of the present study indicate that the rate of resistance to beta-lactams, sulfonamides, tetracyclines, aminoglycosides and fluoroquinolones in ESBLs-producing K. pneumoniae isolates is high in Zahedan and colistin sulfate followed by imipenem and amikacin are the most effective drugs for treatment of infections caused by these isolates. These findings could be used to improve the treatment of infections caused by ESBLs-producing K. pneumoniae in this region.