1. Introduction
Hearing loss is one of the most common sensory disorders that can significantly affected quality of life [1]. Syndromic and non-syndromic hearing loss display in autosomal dominant, autosomal recessive, Y-linked, X-linked or mitochondrial pattern of inheritance [2]. Autosomal recessive nonsyndromic hearing loss (ARNSHL) accounts for up to 80% of cases of NSHL [3]. To date, 61 genes and more than 100 genetic loci have been identified in ARNSHL (http://hereditaryhearingloss.org/).
The most frequently genes involved in ARNSHL are those encoding myosin XVA (MYO15A, MIM# 602666), gap junction protein beta 2 (GJB2, MIM# 121011), solute carrier family 26 (anion exchanger) member 4 (SLC26A4, MIM# 605646), transmembrane channel- like 1 (TMC1, MIM# 606706), otoferlin (OTOF, MIM# 603681) and cadherin-related 23 (CDH23, MIM#605516), each of which has been contained more than 20 various mutations that most of them have been detected in consanguineous families [4]. In present study, we report a case of novel mutation discovered from the direct mutation screening of all exons in the MYO15A gene in an Iranian patient with hearing loss disorder (HLD).
2. Case Presentation
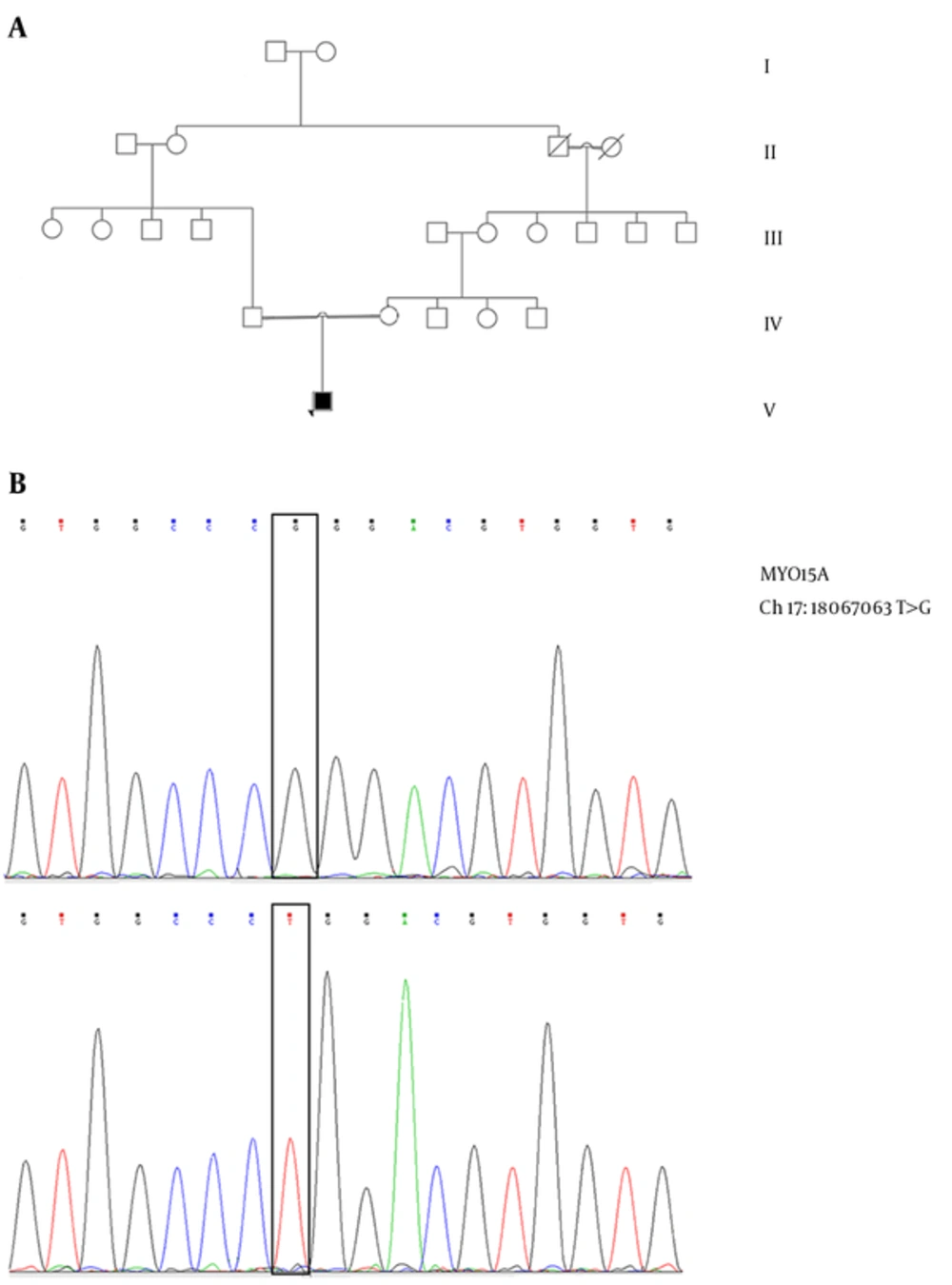
The patient was a 24-years-old male, the only-child of Iranian consanguineous couple (Figure 1A). He was diagnosed with congenital hearing loss and had no dysmorphic features. There was no significant history of hearing loss in this family and no history of systemic disease in the patient.
2.1. Molecular Analysis
Genomic DNA was extracted from peripheral leukocytes of the patient by the standard salting out protocol, and the PCR was conducted under the following conditions: 200 μM deoxyribonucleotide triphosphates (dNTPs), 100 ng genomic DNA, 2.5 units supertaq polymerase, 1.5 mm MgCl, and 25 pmoL each primer (Table 1). Amplification Carried out in 25 μL volumes and 35 cycles: 94°C for 1 minute, 64°C for 35 seconds and 72°C for 45 seconds. Direct sequencing of the 66 exons performed by using the big dye terminator cycle sequencing ready reaction kit on an ABI Prism 3700 automated genetic analyzer. Finally, the sequencing reactions were carried out and the sequences were compared to the reported gene sequence using the BLASTN program.
| Exon | Forward Primers (5' - 3') | Reverse Primer (5' - 3') | Amplicon Size, bp |
|---|---|---|---|
| 2 | Multiple - available upon request | ||
| 3 | ATG ACC AAG CCA GGG GTC | CTC TGG CTG GGA GGG TG | 223 |
| 4 | GAC CCA TGC CAG AAC CAG | AGA AAT CTG TGC GTC CCA CC | 204 |
| 5 | ATC TGT CCG GAT GGA AAC AG | TCT GAC TCA TGG CTC AGG TG | 311 |
| 6 - 7 | GGG AGG TGT GGG AGC TTA G | TCG GGA GTA CAT GAG GTG TG | 499 |
| 8 | TCC TGG AGA GAG TGG TGG TC | CTA GGA CAG GCC TTT GGA TG | 239 |
| 9 - 10 | GGG TGT CCC CAG CTA TGC | TAT CTG TAC CTC CCA CCC CG | 435 |
| 11 | GTT CTC ATC TGC AGC CCA CT | AAA CTC ACC CTC CCC AAA TC | 365 |
| 12 | CAA CTC AGG CCA CCA CAC TA | AAA ACA GGA ACA AGT GAT ATG TGC | 381 |
| 13 | GAC TAC TGG CAT GAG CCA CA | TGA CCC AGG GAC AGA GAG AG | 335 |
| 14 - 15 | GCT TTC CGG AGG CAG AG | GAG GGA GGC GAG ACC TTG | 385 |
| 16 | AGG GAA GGT AGG GGC AAA | CTG TCT CCA AGG AGG TCC AC | 231 |
| 17 | ATT CAA CAT GGG AGG GAG G | TGA GGA CAT GAG GCT GAG AG | 269 |
| 18 | ATA GTG AGG TTG CCA CCA GG | TCT CCA ACA GCT AGC AGC AC | 262 |
| 19 | TCC CTC CTA GGA TAG ACA GAG AG | AAG GCA GGC TGG GTG TG | 212 |
| 20 | TTC CTC CTC ATT TCG GTC TC | CAA GGT CAC ACA GCA TGG G | 441 |
| 21-22 | TTC CTC CTC ATT TCG GTC TC | CAA GGT CAC ACA GCA TGG G | 441 |
| 23 | TAG CAG ACA CCT CGG GTA GG | GAC TCA GTA GTT GTG GAC CCC | 241 |
| 24 | CTT AGT CCA GCC TCC TGG C | TTC AGG CGT GAC CTC TCC | 297 |
| 25 | AGG GCC TCT CTA CCT TTT GG | CTA AGT GCC CTT TCC CCT TC | 219 |
| 26 - 28 | GTG CCG GTC GTC ACC TC | CCC AGG GCA AGG ACA ATG | 569 |
| 29 | CAC AGA GCA GTG GGT CCA G | CTC ATG GCC CAG TTT CAG G | 231 |
| 30 | GGG GAC TGG AAG GAA CAA C | CTT TAA GAC CCT GCC TTG GG | 368 |
| 31 | CAG CCC TCA GCC CCA AG | ACT GGG CCC TGC TGA CTC | 300 |
| 32 | GCA CAG CCA AAC TGG ACT C | CCT TCT GCC TGG GAG TGG | 566 |
| 33 | TCT GTT CAT GTT TAG GGT CTG G | CTC AGC CTG TCC CAG CAG | 396 |
| 34 - 35 | GGA GAA AGC CAC TGA ATA CCA G | GAG AAG CTC TCA GGT CAC CC | 553 |
| 36 - 38 | AGT GTC AGG TGC CTG TTG C | TCC TCT TTA CAG CTT GTG TCT CC | 620 |
| 39 - 40 | TCT GGA GTC CCA GAG AGC AG | GGG CCA TGA TGG ACA CTC | 549 |
| 41 - 42 | ATG TGA TGG GAA AGG GAG AC | CTG TGC CCA CAG ACT TCC TC | 460 |
| 43 | ACT CTA GCC TGG GGG ACA AC | CCC AAG TCC TAG ACC CTC CT | 320 |
| 44 | CCC AGG AGG ACA GAA AAA GG | GGG AGG GGG AGA TTC AAT AA | 356 |
| 45 | AGT ATA GTC CAG CCT GGG TCC | CTG GCT GTG CCT CTG ACT G | 202 |
| 46 | TGG CCA TCT CAT CCA TTT CT | CAC AGC TAG GAG CTG CAC AC | 397 |
| 47 | GAA CCA GCT GGA CAC ACA GA | AAA TGG GTT TGC TTC AAT GG | 458 |
| 48 | GGG CAG GAC AGG ATC AGA AG | AGG GAG ATC CCT GTT GCT G | 291 |
| 49 - 50 | CTA GGC CTC TGG GAG TGG | CAC CAC GAG TGG GTG AAA C | 400 |
| 51 | CCC CTT AGT CAC AAG ACA AGA C | TTA TCC CCA CTC GCC TCA C | 319 |
| 52 | CTA GGG GTT CGC TTG TCA GT | AGT GGG GCC TCC GAG ACT | 295 |
| 53 | TGT GAG GCT CAT TTC AGT GC | AGG GTG CTG AGA ATC AGA GG | 352 |
| 54 - 55 | TGT GTC CCC TTT CTG TTC TG | TGA TAG ATG GGG AAA CTG AAC C | 534 |
| 56 | GTG CCC ACC CTG TTC TTA TG | CCT CCT GGA GCA TGG ACA C | 222 |
| 57 | TCT CAG CTC AAT CCC AGG AG | TCC ACC CAG TCC CCA AG | 271 |
| 58 | ATG GGG GAG TAA ATG CCT TC | GGC TTG TGT CTC CCA TTC AT | 594 |
| 59 | CAG GAG ACA AGG GCT GTC C | CTG GAG CCT GGG CTG TC | 214 |
| 60 | AGA AGG ACA GAG GTC AAG CC | AAA TCT GGG TGG AGG GC | 236 |
| 61 | AAG CTG TGT CCC AGA ACA GG | ACA GGG CCT GAA TCA TGA AC | 418 |
| 62 | TGA GAG GGC AGG GTT GC | CAT GCA TGT CCC CAG GTC | 271 |
| 63 | ACA GTG AGG ATT GCC TGA GC | TAC CCA TCC TCC ATG ACC AC | 269 |
| 64 | AGC CCA GAG AAG CTA TGC AG | AGG CTC AGA GGA GGG AAG AG | 374 |
| 65 | TGG TTG AGA CTA TCC TCG CC | GAC CTG ACC TAT CTT GGA GCC | 271 |
| 66 | CAA GGT AAG AGC TGG GGA AG | TTG ATC CTG AGA GGT TCA GTG | 240 |
The effect of Candidate variant in protein structure and phylogenetic conservation was predicted by using bioinformatic tools like PolyPhen-2 (PolymorphismPhenotyping v2).
3. Results
Sequencing analysis of the patient, after comparison with MYO15A reference sequence in 1000 Genomes database, demonstrated a novel mutation, a homozygous missense mutation, c.9698T > G in exon 59 of the MYO15A gene (Figure 1B). The c.9698T > G mutation is novel and has not been previously described in HLD.
The novel homozygous missense mutation was predicted to be possibly damaging by in silico prediction of the recognized variant, Polyphen 2 (probably damaging, score 1.00).
4. Discussion
We analyzed an affected person with HLD with PCR and direct sequencing of coding exons of the MYO15A gene. As a result we identified a genetic variant of MYO15A in ARNSHL patient. The mutation identified in our patient involved a novel homozygous mutation, c.9698T > G in exon 59, which results in substituting a Leusin to an Arginin (p.L3233A) in the ferm domain and tail region of the Myosin protein. So, this exchange amino acid results in alter a nonpolar amino acid to polar positively charged that can modify interaction of tail region of myosin with membranous compartments and change its movement to actin filaments. This change is predicted to negatively affect the usual function of the myosin XVa protein by in silico. Overall, it seems that this amino acid has an important role in the myosin-XV protein, and mutation at this site results in pathogenicity and deafness.
MYO15A has 66 exons and its coding protein, myosin XVa, has a critical role in formation of stereocilia in hair cells of the cochlea [5]. Myosin XVa in the organ of Corti is localized completely at the tips of stereocilia and is anactin-activated ATPase that uses the hydrolysis of ATP to move on actin filaments. The tip of a stereocilium is one of the proposed sites of mechano-electrical transduction and the site of stereocilia growth [6]. Myosin XVa is required for proper function and formation of the mechanotransduction machinery. All myosins are composed of one or two heavy chains and several light chain. The tails of the myosins presumed to bind to membranous compartments, which would be moved relative to actin filaments [7].
Twenty-nine mutations have been described in MYO15A in HGMD (http:// www.hgmd.org).
In summary, this is the first case with hearing loss in south-west of Iran confirmed by genetic analysis involving a novel MYO15A gene mutation. Further studies are required to understand the structural and functional changes of proteins involved in this disorder and their relations with phenotypic spectrum. Genotype-phenotype relations of MYO15A mutations and degree of hearing loss suggest that mutations in all 66 exons cause intense deafness.