1. Background
Staphylococcus aureus is a Gram-positive, facultative anaerobic non-spore-forming, catalase-positive bacterium [1]. Staphylococci, especially Staphylococcus epidermidis, are part of the normal flora of human skin, and gastrointestinal and respiratory tracts. S. aureus is the most significant pathogen of human infections, mainly wound, respiratory tract and skin infections that is associated with several pathological processes such as food poisoning, pneumonia, bacteremia, folliculitis, and osteomyelitis [2-4]. This organism is one of the most important causes of nosocomial infections, which become increasingly more resistant against the antibiotics of beta-lactam and vancomycin groups [5, 6]. Coagulase is an enzyme produced by S. aureus that causes clotting of blood in the human host. S. aureus secretes two forms of coagulase enzyme, bound coagulase and free coagulase [7, 8]. Free coagulase binds with coagulase-reacting factor (CRF) in plasma and creates a complex, staphylothrombin [9-11]. Dawnson et al. Conducted a descriptive study in China on specific molecular and epidemiological characteristics of S. aureus in patients in Xinyang, China [12]. The Coa gene, coding for the coagulase enzyme, can be used for DNA-based diagnosis of S. aureus. The coa gene is highly polymorphic because of differences in the sequence of the 30 variable region. Analysis of the Coa gene in a variety of staphylococcal species has shown diversity in the amino acid sequence and the number of tandem repeats at the 30 end. There is heterogeneity in this domain, including the number of 81-bp tandem short sequence repeats encoding repeated 27 amino acid sequences in the C-terminal region. Detecting the coagulase enzyme in staphylococci infections is important because it is considered as one of the pathogenic factors of this bacterium [13, 14]. Studies on molecular diagnosis of genes encoding coagulase of S. aureus using different techniques such as PCR and RFLP can be conducted. PCR, based on molecular methods, is often preferred to diagnose antibiotic resistant genes. Rapid and reliable methods for antibiotic susceptibility are important to institute appropriate therapy [13-15]. Accordingly, the present study aims to determine the antibiotic resistance patterns and their relationship with presence of coagulase gene in clinical isolated of S. aureus collected from hospitals of Nowshahr and Chalous.
2. Methods
2.1. Sample Collection
In the current study, that is experimental study, 30 isolates of staphylococci, including 6 samples of S. epidermidis, 4 samples of S. saprophyticus, and 20 samples of S. aureus, which were obtained from hospital clinical samples (pharynx, nose, urine, trachea, lung, vagina, joint fluid, and lesion) of hospitals in Nowshahr and Chalous, between March 2015 to august 2015. It should also noteworthy that ethical issues regarding collecting the samples above were thoroughly regarded in that the researchers were permitted to gather the required samples. All the samples were cultured and purified in blood agar media.
2.2. Species Identification
Using microbial standard methods including, catalase, coagulase (tube and slide), mannitol fermentation on mannitol salt agar, brad parker agar, DNase agar, the isolated S. aureus was confirm. Also for detecting coagulase-negative staphylococci (epidermidis and saprophyticus) from micrococcus Bacitracin test was used. In order to distinguish S. epidermidis and S. saprophyticus, Novobiocin resistance test was used. Isolates were stored at -80°C in Nutrient Broth (Merk-Germany) supplemented with 30 percent Glycerol
2.3. Susceptibility Testing
The sensivity of isolates (S. aureus, S. epidermidis, S. saprophyticus, and micrococcus) was examined by disk diffusion method based on CLSI guide direction, including: penicillin, oxacillin, gentamicin, chloramphenicol, vancomycin, kanamycin, and nalidixic acid.
2.4. DNA Extraction
To extract nucleic acid from bacteria, the boiling method proposed in a study by Hosseinpour et al. was used [11]. DNA-containing micro-tubes were kept at -20°C.
2.5. PCR Technique
Isolates were tested for the presence of coa gene using two sequence of primers as described by Hookey et al. [16]. Including the Forward primers (5-ATA-GAG-ATG-CTG-GTA-CAG-G-3) and Reverse (5- GCT-TCC-GAT-TGT-TCG-ATG-C-3). Each of extracted 50-microliter tube of PCR reaction contained 2.5 microliter nucleic acid, a master mix of 3.2 µL, forward primer of 0/8 µL, reverse primer of 0/8 µL, and 12.7 µL of distilled water. Reaction was performed in the thermo cycler ( Eppendorf- Germany) by denaturation at 95°C for 5 minutes, followed by 40 cycles of 95°C for 30 seconds, 55°C for 1 minute and 72°C for 1 minute. Final extension was 7 minutes at 72°C in the end of cycle. The products of the PCR were analyzed by electrophoresis in a 2% agarose gel.
Treatments were examined in three intervals and the results were analyzed using one-way analysis of variance (ANOVA), univariate ANOVA, and t-test at the probability level of P < 0.05. Moreover, the comparison of means was presented using Duncan’s multiple range test (P < 0.05). All the statistical analyses were carried out using SPSS (V. 16) and graphs were drawn using Microsoft Excel 2007.
3. Results
3.1. Antibiotic Resistance in Isolates of S. aureus, S. Epidermidis, and S. Saprophyticus
Table 1 shows that coagulase-positive strains are more resistant to antibiotics compared with coagulase-negative strains.
| Antibiotics | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | K | V | PG | NA | GM | OX | C | TE |
| CoA+ | 25 | 0 | 100 | 55 | 20 | 55 | 0 | 20 |
| CoA- | 0 | 0 | 100 | 50 | 10 | 10 | 0 | 10 |
Abbreviations: C, chloramphenicol; GM, gentamicin; K, kanamycin; NA, nalidixic acid; OX, oxacillin; PG, penicillin; TE, tetracycline; V, vancomycin.
aValues are expressed as %.
Coagulase-negative strains show 10% resistance to nalidixic acid, oxacillin, and tetracycline, they were sensitive to kanamycin, vancomycin, and chloramphenicol.
The results reveal that coagulase-positive staphylococci samples were 100% sensitive or semi-sensitive to chloramphenicol, 100% sensitive to vancomycin, 45% sensitive to oxacillin, 80% sensitive to tetracycline, 75% sensitive to kanamycin, 45% sensitive to nalidixic acid, and 80% sensitive to gentamicin. All of the isolates were resistant to penicillin.
Also, the results show that coagulase-negative staphylococci were 90% sensitive or semi-sensitive to gentamicin, and 100% sensitive or semi-sensitive to kanamycin and vancomycin. All of the isolates were resistant to penicillin, and 90% sensitive to oxacillin.
3.2. Polymerase Chain Reaction (PCR) for Coagulase Gene in S. aureus Samples
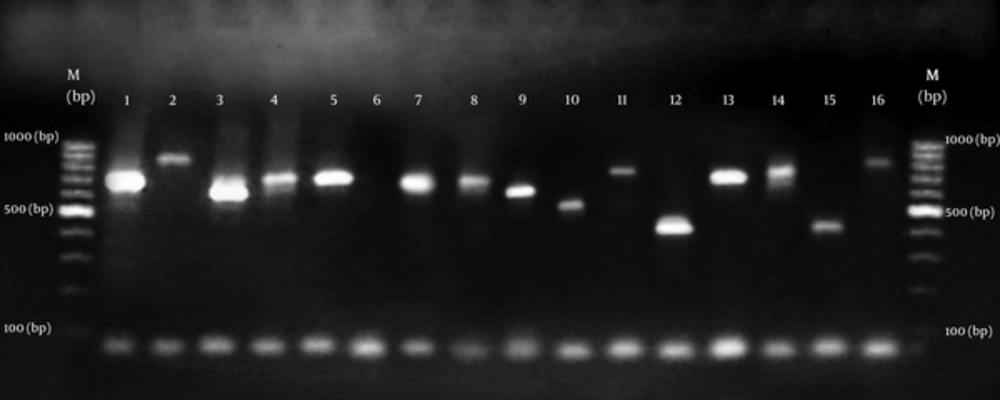
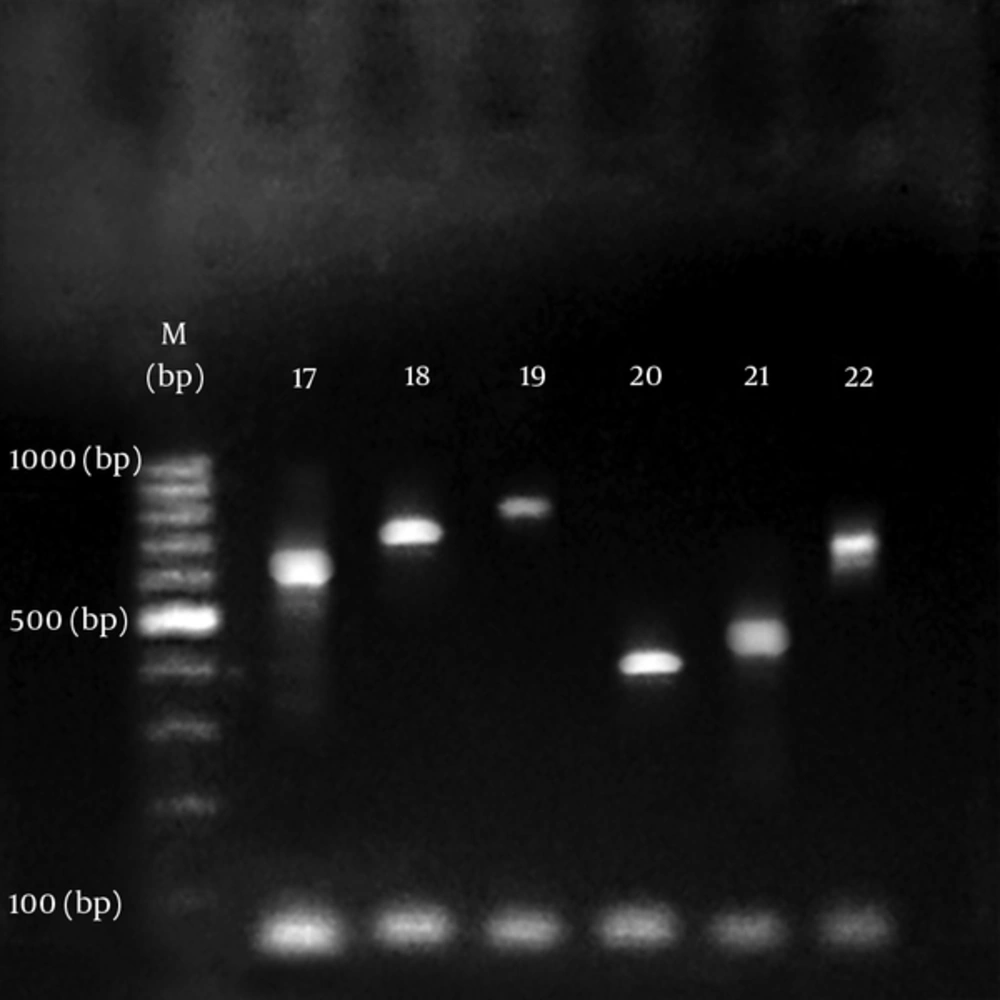
All isolates of S. aureus produced PCR amplicon with the coa primers. The agarose gel analysis of the digestion products showed 15 diferent sizes ranging from approximately 400 to 900 bp. As summarized in Figures 1 and 2. There was no amplification on product of DNA from s. epidermidis and s. saprophyticus. Results show that S. aureus samples in biochemical tests contained coagulase enzyme as well as containing it in PCR test (Table 2).
(1 -16 isolates). Molecular size marker. Lane 1 (700 bp); Lane 2 (890 bp); lane3 (610 bp); lane 4 (710 bp); lane 5 (710 bp); lane 6 (Negative control); lane 7 (700 bp); lane 8 (700 bp); lane 9 (630 bp); lane 10 (530 bp); lane 11 (790 bp); lane 12 (410 bp); lane 13 (710 bp); lane 14 (735 bp); lane 15 (420 bp); lane 16 (816 bp).
| Coagulase | ||
|---|---|---|
| Antibiotics | Coagulase + | Coagulase - |
| Oxacillin | 4/487 ± 19/090cd | 16/700 ± 2/830ab |
| Chloramphenicol | 4/442 ± 23/950b | 15/500 ± 3/028a |
| Tetracycline | 11/815 ± 29/220a | 17/000 ± 2/828ab |
| Vancomycin | 2/745 ± 20/800bc | 16/000 ± 1/333abc |
| Kanamycin | 4/125 ± 18/530cd | 1/729 ± 15/900abc |
| Nalidixic acid | 5/026 ± 15/210d | 2/936 ± 13/200c |
| Gentamicin | 4/651 ± 20/590bc | 4/111 ± 15/300bc |
| Penicillin | 3/976 ± 15/150d | 3/551 ± 13/110c |
aNumbers with common words in every column do not have significance difference with each other statistical P < 0.05.
4. Discussion
Staphylococcus aureus is the most important pathogen of human resources mainly wound, respiratory tract and skin infections. Infection spreads in hospitals because of close contact and the point that hospitals staff and patients carry antibiotic resistant S. aureus specially (MRSA) in their nose or on their skin [3, 4, 17].
Staphylococci, especially S. epidermidis are parts of the natural flora of human skin and digestive and respiratory tracts. [18].
This organism is one of the main agents of nosocomial infections which are becoming increasingly resistance to antibiotics of beta-lactam and vancomycin groups [5]. The main factor in the survival of these microorganisms in spite of numerous anti-staphylococcal antibiotics is their ability to become resistant against antimicrobial agents.
Conducting antimicrobial tests on pathogenic bacteria in patients can provide good guidance regarding how to proceed for treatment. Although various studies have shown different results of bacterial resistance and carriers which may be related to various bacterial detecting methods [17]. Morelli et al. in a study on 1,633 unique S. aureus isolates reported. All isolates were susceptible to trimethoprim sulfamethoxazole, linezolid, ceftaroline, and rifampin, while the majority of isolates were susceptible to clindamycin, tetracycline, and mupirocin [19]. Also, Sun et al. conducted a descriptive study in China on specific molecular and epidemiological characteristics of S. aureus in patients in Xinyang, China. Several antibiotic-resistant genes were examined. All the samples were sensitive to vancomycin and daptomycin, 98.1% resistant to penicillin, and 9.3 resistant to chloramphenicol [12].
Coagulase protein is a main virulence factor in S. aureus so addition to its important role in diagnosis S. aureus [17, 20].
The coa gene amplificationhas been considered a simple and accurate method for typing of S. aureus isolated from distinct sources, the coagulase protein is an important virulence factor of S. aureus. Like spa, coa has a polymorphic repeat region that can be used for differentiating S. aureus isolates. The variable region of coa gene is comprised of 81 bp tandem short sequence repeats (SSRs) [21].
In the current study, 20 samples of S. aureus and 10 samples of coagulase-negative staphylococci were studied to antibiotic resistance patterns against disks of penicillin, tetracycline, gentamicin, chloramphenicol, oxacillin, vancomycin, kanamycin, and nalidixic acid. Then, all of the isolates were analyzed for the presence of coagulase gene using polymerase chain reaction.
As to the results, coagulase-positive samples showed higher resistance towards antibiotics. Statistically, there was not a significant difference between inhibition zone diameters of oxacillin, nalidixic acid, gentamicin, kanamycin, and penicillin. Isolates of S. aureus were 25% resistant to kanamycin and 55% to nalidixic acid. Resistance to tetracycline and gentamicin was 20%. All of the isolates were sensitive to vancomycin and resistant to penicillin.
Staphylococcus epidermidis and S. saprophyticus isolates showed 10% resistance to oxacillin, tetracycline, and kanamycin. All of the isolates were sensitive to chloramphenicol and vancomycin.
To conclude, prevalence of S. aureus and S. epidermidis in hospitals is increasing. Coagulase-positive samples showed more resistance to antibiotics, which confirms the virulence of S. aureus based on Talebi Satlou et al. study [14]. The reports in this study and other reports emphasized that they were 100% resistant to penicillin. Coagulase gene has variance of amplicon which is in line with the reports of Talebi Satlou et al. [14]. In the present study, PCR products of different sizes were obtained by amplification of the coa gene, similar to other researchers in Iran and different parts of world [16, 17, 22].
Results of the study suggest that PCR is a safe and accurate way to detect coagulase gene in bacteria. Thus, choosing the right antibiotic, continuous monitoring of resistance patterns, and proper treatment of staphylococcal infections are of most importance. As to the literature regarding PCR technique, which has been carried out with other methods such as Ethanol, various ranges of size have been observed, highlighting such variety for Coagulase gene [22, 23]. Genotypic variation among different S. aureus isolates, which may be considered as an important criterion when treating staphylococcal infection [24].
4.1. Conclusions
Coagulase gene polymorphism analysis provides easy and early analysis of the clonal relatedness of isolates. This study emphasize the genotypic variation among different S. aureus isolates, which may be considered as an important criterion when treating staphylococcal infections. And finally, since coagulase gene has a variety of band sizes and, as reported in different sources, is polymorphic; more studies are required in this area. Moreover, due to the importance of staphylococci, genomic studies on enterotoxins in S. aureus and coagulase-negative staphylococci is suggested.