1. Background
Pseudomonas aeruginosa is a Gram-negative, glucose non-fermentative bacillus that is often involved in nosocomial infection outbreaks (1). This opportunistic pathogen can cause a variety of infections including pneumonia, septicemia, meningitis, urinary tract infections, and burn wound infections (2, 3). The antibiotic therapy of nosocomial infection caused by P. aeruginosa is often complicated because the drug resistance of this organism has increased recently (4). The terms pan-drug-resistant (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) are defined as resistance to all antibiotics in all different categories of antimicrobial agents, resistance to at least one antibiotic in all except one or two antimicrobial categories, and resistance to at least one antibiotic in ≥ 3 antimicrobial categories, respectively. The antimicrobial categories contain aminoglycosides, carbapenems, cephalosporins, fluoroquinolones, penicillins, monobactams, and polymyxins (5). Carbapenems are effective agents for the treatment of important Gram-negative bacilli infections, especially P. aeruginosa (3). Among various mechanisms involved in resistance to carbapenems in P. aeruginosa, the production of metallo-beta-lactamases (MBLs) is a very important mechanism through which all carbapenems are hydrolyzed (4, 6, 7). Furthermore, the metallo-beta-lactamase genes located on integrons may elucidate their wide distribution between strains (8). At present, different groups of MBLs have been identified in P. aeruginosa but the IMP (Imipenenase) and VIM (Verona integron-encoded metallo-beta-lactamase) groups are the most widespread groups of MBLs (9-). This study was done to detect the antimicrobial susceptibility profile and the rate of MDR, XDR, and PDR among P. aeruginosa isolated from multiple hospitals of Qom province and to determine the distribution of MBL genes among carbapenem non-susceptible isolates.
2. Methods
2.1. Bacterial Isolation and Identification
In a study that was done between November 2013 and October 2014, 200 non-duplicated P. aeruginosa isolates were collected from different specimens at Shahid Beheshti, Nekoei, Kamkar, and Valiasr Hospitals (affiliated to Qom University of Medical Sciences, Iran). The clinical specimens included tracheal aspirate, urine, burn wound, blood, surgical wound, and stool. The samples were immediately inoculated in MacConkey agars. The identification of the isolates was carried out using standard microbiological and biochemical methods including Gram-staining, colony morphology, glucose oxidation, citrate utilization, oxidase test, and pigment production (13).
2.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility profile of isolates was tested by standard disc diffusion method according to the Clinical Laboratory Standard Institute (CLSI) guidelines (14). The antibiotic discs contained ceftriaxone (30 µg), ceftazidime (30 µg), ceftizoxime (30 µg), cefotaxime (30 µg), gentamicin (10 µg), Amikacin (30 µg), ciprofloxacin (5 µg), piperacillin (100 µg), piperacillin-tazobactam (100/10 µg), imipenem (10 µg), meropenem (10 µg), Aztreonam (30 µg), colistin (10 µg), and polymyxin B (300 units) (MAST, United Kingdom). Escherichia coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as control strains in the tests. The isolates that were intermediate-resistant to antibiotics were considered as resistance. The minimum inhibitory concentrations (MIC) of all imipenem non-susceptible isolates for imipenem was obtained by microbroth dilution method according to the recommendations of the standard protocol of CLSI (14).
2.3. Detection of MBL Producing Isolates and MBL Genes
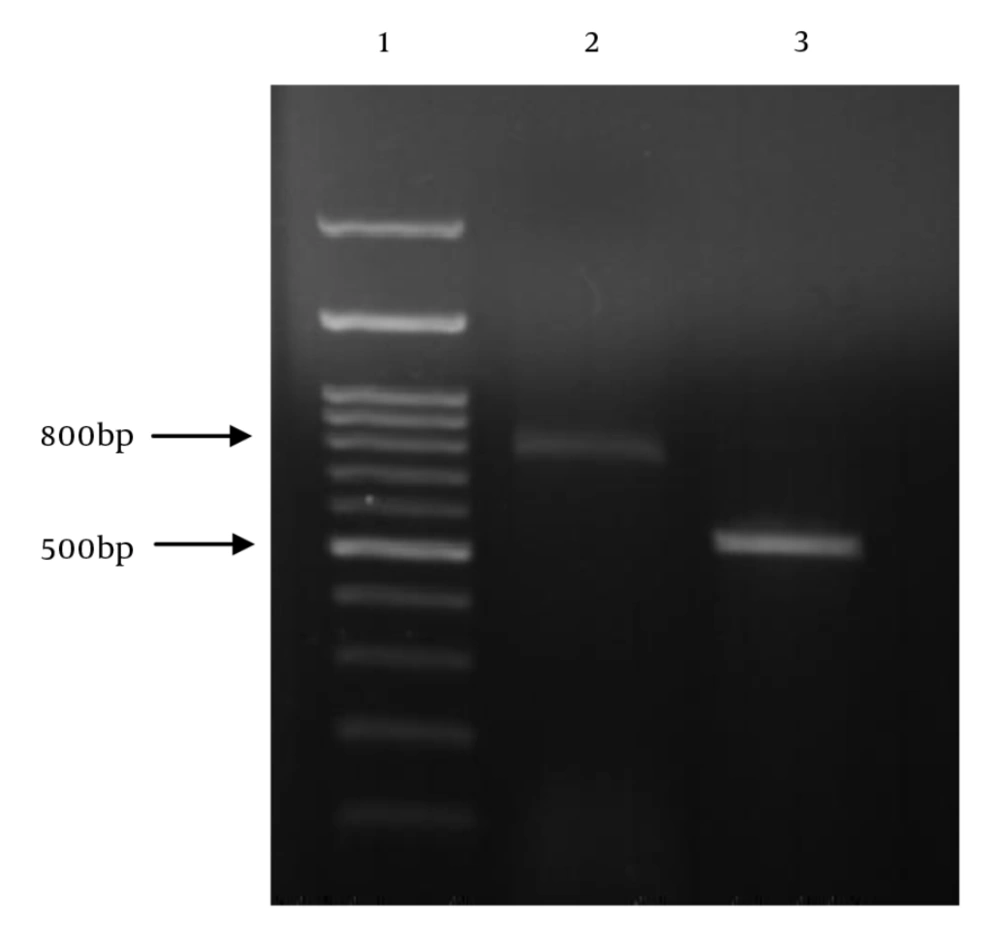
The detection of MBL production in carbapenem-resistant isolates of P. aeruginosa was carried out by double disk synergy test (DDST) (15). DNA of carbapenem resistant isolates was extracted by boiling method (15). The detection of blaIMP and blaVIM genes in P. aeruginosa isolates was achieved by PCR with specific primers (Table 1) (16). The amplification steps were as follows: Initial denaturation for 3 minutes at 94ºC; 35 cycles with denaturation for 1 minute at 94ºC, annealing for 1 minute at 53ºC, extension for 1 minute at 72ºC, and final extension for 7 minutes at 72ºC. Gel electrophoresis was performed in 1.2% agarose gel at 85 V. P. aeruginosa ATCC 27853 was used as the negative control strain and P. aeruginosa harboring blaIMP and blaVIM genes (obtained from Dr. Hashemi, Iran) were used as the positive control.
| Primers | Sequence (5'- 3') | Product Size, bp |
|---|---|---|
| IMP- F | ACCGCAGCAGAGTCTTTGCC | 587 |
| IMP- R | ACAACCAGTTTTGCCTTACC | |
| VIM - F | ATGTTCAAACTTTTGAGTAAG | 801 |
| VIM- R | CTACTCAACGACTGAGCG |
The Sequence of Primers Used for Detection of MBL-Encoding Genes
2.4. Statistical Analysis
Differences in variables were analyzed using Chi-Square test and P-values of less than 0.05 were considered statistically significant. All the available data were analyzed by a computer program (SPSS).
3. Results
In total, 200 P. aeruginosa isolates were collected from tracheal aspirate (78 isolates, 39%), urine (52 isolates, 26%), burn wound (42 isolates, 21%), blood (13 isolates, 6.5%), surgical wound (12 isolates, 6%), and stool (three isolates, 1.5%). 73 patients (73%) were male and 26 (27%) were female. The age range of the patients was 20 - 86 years. P. aeruginosa isolates were recovered from several hospital settings as follows: intensive care unit (ICU) (39%), burn unit (21%), trauma and emergency wards (16.5%), general surgery wards (14%), and internal wards (9.5%). Among the P. aeruginosa isolates, 76 isolates (38%) were multidrug-resistant (MDR), 52 isolates (26%) were pan-drug-resistant (PDR), and 10 isolates (5%) were extensive drug resistant (XDR) (Table 2). On the other hand, among the 200 P. aeruginosa isolates, 104 isolates (52%) were resistant to imipenem and meropenem. The majority of isolates (94.4%) showed MIC ≥ 32 mg/L for imipenem. The results of the DDST method showed that among 104 carbapenem non-susceptible isolates of P. aeruginosa, 40 isolates (38.4%) produced metallo-beta-lactamases. The P. aeruginosa isolates producing metallo-beta-lactamases were mainly obtained from tracheal aspirate (63%), burn wound (28%), and urine (9%). Of the 104 carbapenem non-susceptible isolates, 38 (36.5%) isolates carried the blaIMP gene and two (1.9%) isolates carried the blaVIM gene (Figure 1). Of the 40 MBL-producing isolates, 21 isolates (52.5%) were PDR and 19 (47.5%) isolates were MDR. The Pearson Chi-Square analysis showed that the rates of resistance to imipenem, meropenem, ceftazidime, cefotaxime, gentamicin, amikacin, ciprofloxacin, and piperacillin-tazobactam were significantly higher in isolates with blaIMP gene than in isolates without the gene (P-values ≤ 0.001). The rates of resistance to piperacillin and colistin were statistically higher in isolates with blaIMP gene than in isolates without the gen (P-values ≤ 0.05). The rates of resistance to tested antibiotics were not significantly different between isolates in which blaVIM gene was detected and isolates in which it was not detected.
| Antimicrobial Agent | Susceptible, % | Intermediate, % | Resistant, % |
|---|---|---|---|
| Ceftizoxim | 7 | 11 | 82 |
| Cefotaxim | 7 | 28 | 65 |
| Imipenem | 48 | 7 | 45 |
| Meropenem | 48 | 10 | 42 |
| Ceftazidime | 41 | 9 | 50 |
| Ceftriaxone | 22 | 28 | 50 |
| Ciprofloxacin | 41 | 13 | 46 |
| Gentamicin | 37 | 17 | 46 |
| Aztreonam | 34 | 23 | 43 |
| Piperacillin | 59 | 6 | 35 |
| Piperacillin-tazobactam | 49 | 23 | 28 |
| Amikacin | 63 | 9 | 28 |
| Polymyxin B | 97 | - | 3 |
| Colistin | 88 | - | 12 |
Antimicrobial Susceptibility Patterns of Pseudomonas aeruginosa Isolated from Four University Hospitals in Qom, Iran
4. Discussion
P. aeruginosa is one of the most important Gram-negative opportunistic pathogens, which is recognized as an agent of nosocomial infections. Our study showed that 39% of the P. aeruginosa isolates were collected from patients in ICUs, indicating that invasive devices play an important role in P. aeruginosa infections (17-20). The rate of resistance to carbapenems in P. aeruginosa isolates in this study was 52%, which is higher than the rates of resistant to carbapenems (32% and 12.4% among P. aeruginosa isolates) in previous studies reported from Iran (21, 22). These results indicate that carbapenem-resistant P. aeruginosa has increased recently (17, 23). In the present study, the distribution of MDR, PDR, and XDR was found to be 38%, 26%, and 5%, and this finding is the first report of the distribution of PDR and XDR isolates in P. aeruginosa from Iran. In the present study, higher rates of resistance to third-generation cephalosporins including ceftizoxime (93%), cefotaxime (93%), ceftazidime (59%), and ceftriaxone (78%) were observed, implying that third-generation cephalosporins are no longer secure to treat P. aeruginosa in our hospitals. Therefore, finding an appropriate therapy for infections caused by multidrug-resistant P. aeruginosa is a newly difficult situation. Our results were consistent with the observations in previous studies from Iran indicating that the majority of P. aeruginosa isolates showed MIC ≥ 32 mg/L for imipenem (21). In our study, we tested MBLs production by using the DDST method and demonstrated that 38.4% (n = 40) of the carbapenem non-susceptible P. aeruginosa isolates were MBL positive. This result is consistent with those of previous reports from Iran and Brazil (21, 24, 25). In the present study, 61.6% of the carbapenem non-susceptible P. aeruginosa were MBL negative, which suggests that other factors such as deficiency in porin or reduced expression of outer membranes proteins might be associated with carbapenem resistance. In the present study, the distribution of IMP-type MBLs among carbapenem-resistant isolates of P. aeruginosa was 36.5%. Other studies demonstrated that 5.77% of P. aeruginosa isolates from the northwest of Iran (2010) (21) and 24% from the center of Iran (2013) (26) were IMP-type positive. These results indicate that the distribution of IMP-type MBLs among the isolates of P. aeruginosa in Iran in recent years has increased significantly. In our study, 1.9% of the imipenem non-susceptible isolates of P. aeruginosa carried VIM-type MBLs. Bagheri Bejestani et al. reported that blaVIM gene was not detected among the studied isolates in the center of Iran (27). However, several previous reports revealed that the genes of blaIMP and blaVIM-type MBL are often encoded on mobile genetic cassettes, which can easily spread among isolates.
4.1. Conclusions
This study showed that susceptibility to carbapenems among isolates of P. aeruginosa reduced in Iran and the IMP-type MBLs were the most prevalent MBLs among imipenem non-susceptible P. aeruginosa isolates in this area. MBL-producing P. aeruginosa in recent years has become a serious problem. The rapid identification of MBL-producing isolates is very important and the use of specific methods is necessary to control the further transmission of infection by these organisms.