1. Background
Hepatitis B is a viral infection and a major cause of viral hepatitis, cirrhosis and hepatocellular carcinoma in different regions of Asia and Africa [1]. According to WHO reports just over 2012 about 2 billion people have been infected with HBV and more than 240 millions suffer from chronic infection whilst, 600,000 pass away from acute and chronic infection per year [2]. HBV is a member of Hepadnaviridae and is classified into 8 genotypes from A to H in accordance with at least 8% difference in complete nucleotide sequences or more than 4% in S gene [3, 4]. Though, some researchers have categorized HBV into 10 genotypes (A to J) [5-8]. As in case of genotypes A to F considerable data is available concerning their geographic distribution, differential sequences and frequency of strain spread [9] in contrast to genotype G which, in spite of efforts by Kato et al. yet no significant information is presented regarding its epidemiology and incidence. So far, only one case of genotype G has been documented from France, Germany and USA [10, 11].
Furthermore, few isolates of genotypes H and F are related and show similar geographic distribution [9]. Although, all genotypes of HBV induce infection in liver but demonstrate different clinical features and potency [12-14]. Patients infected by genotype C or D indicated more mutations in promoter region of nucleus and in comparison with A and B genotypes show less effective response to interferon therapy hence, spread of infection toward cirrhosis and hepatocellular carcinoma is accelerated [15].
In Sunbul et al. study in Turkey, genotype D was detected in 88.7% patients [16]. As Iran is located in Mediterranean region, it is assumed that genotype D is dominant [17]. Eftekhari et al. have reported genotype D the only existing genotype in Iran and other genotypes have not been described however, their study was just restricted to Tehran and Fars provinces and which is not extendible to other parts of the country [18].
2. Objectives
Different genotypes of HBV have different clinical manifestations hence; the objective of this study was to determine HBV genotype in infected individuals in Sistan and Baluchestan (south-east Iran) province.
3. Patients and Methods
In this descriptive-analytical study, we chose all patients who were referred to hepatitis clinic in Zahedan blood transfusion centre from March to August 2012 once their written consents were taken. The inclusive criteria for sera samples were: positive HBeAg as well as positive HBsAg, the latter was proved by ELISA kit (Siemens from Germany) and viral load of more than 10,000 copies/mL. The number of samples selected was 163 and since transmission of HBV in this province is mainly congenital and presence of one genotype in a family is shared with other members therefore, from each family only one member was included in our investigation. In case any sample was HBsAg positive but at the same time anti-HDV, anti-HCV and anti-HIV were also positive, was excluded from our study. Moreover, if any sample had viral load less than 10,000 copies/mL as well as HBeAg negative was expelled too. The selected sera were then incubated at -43ºC followed by DNA isolation by a specific kit manufactured by Roche (Germany), the extracted DNAs were stored at -22ºC for further assays.
The purified DNAs were then subjected to multiplex PCR in addition to exact primers of different genotypes of HBV with specified sequences, as described formerly [9]. All PCR cocktails had final volume of 20 µL after utilization of Master Mix (of Genet Bio company) consisting Taq DNA polymerase 1 unit/10 µL, primers 4 mM, KCL 80 mM, Tris-HCL 20 mM, MgCl2, enzyme stabilizer, sediment, loading dye, pH = 9, 0.5 mM of each as well as dATP (Deoxycytidine triphosphate), dCTP (Deoxycytidine triphosphate), dGTP (Deoxycytidine triphosphate), dTTP (Deoxycytidine triphosphate). Moreover, to Master Mix 1.2 µL of extracted DNA plus 0.5 µL of primers (six pairs) were added and through addition of distilled water the final volume was equal to 20 µL.
PCR programming was based on protocol introduced by Kirschberg et al. [9] with some modification as following. The cycling conditions were: an initial predenaturation at 95ºC for 15 minutes, subsequently denaturation 94ºC for 1 minute, annealing at 60ºC for 1 minutes, an extension at 72ºC for 1 minute for 40 cycles besides a final extension at 72ºC for 10 minutes (Table 1). Once, PCR cycles were completed the product was subjected onto electrophoresis on 2.5% agarose gel. The gel was then stained by ethidium bromide and the results adjacent to DNA ladder (50 bp) were looked into under UV light. However, primer sequences and sizes of each PCR product corresponding to different genotypes are represented in Table 1.
| Genotype and Primers | Sequence | Size of Amplification, bp |
|---|---|---|
| A | 370 | |
| Sens (nt 2331 - 2360) | 5-CGGAAACTACTGTTGTTAGACGACGGGAC-3 | |
| Anti-sens (nt 2701 - 2665) | 5-AATTCCTTTGTCTAAGGGCAAATATTTAGTGTGGG-3 | |
| B | 190 | |
| Sens (nt 1470 - 1491) | 5-CCGCTTGGGGCTCTACCGCCCG-3 | |
| Anti-sens (nt 1660 - 1633) | 5-CTCTTATGCAAGACCTTGGGCAGGTTCC-3 | |
| C | 701 | |
| Sens (nt 2706 - 2741) | 5-CCTGAACATGCAGTTAATCATTACTTCAAAACTAGG-3 | |
| Anti-sens (nt 192 - 165) | 5-AGCAGGGGTCCTAGGAATCCTGATGTTG-3 | |
| D | 147 | |
| Sens (nt 2843 - 2870) | 5-ACAGCATGGGGCAGAATCTTTCCACCAG-3 | |
| Anti-sens (nt 2990 - 2966) | 5-CCTACCTTGTTGGCGTCTGGCCAGG-3 | |
| E | 787 | |
| Sens (nt 2093 - 2122) | 5-CTAATGACTCTAGCTACCTGGGTGGGTGTA-3 | |
| Anti-sens (nt 2880 - 2853) | 5-CCATTCGAGAGGGACCGTCCAAGAAAGC-3 | |
| F | 481 | |
| Sens (nt 2843 - 2871) | 5-ACAGCATGGGAGCACCTCTCTCAACGACA-3 | |
| Anti-sens (nt 109 - 83) | 5-AGAGGCAATAGTCGGAGCAGGGTTCTG-3 |
Primer Sequences and Sizes of the Products
3.1. Statistical Analysis
All statistical analyses were performed using SPSS-15 software, genotypes frequencies were completed by means of cross-tab and χ2 analysis. P value < 0.05 is considered as level of significance.
4. Results
In this study 163 samples, from individuals infected by HBV, were collected including 117 (71.8%) males and 46 (28.2%) females. The average age of victims was 35 ± 13 years, all sera were HBsAg and HBeAg positive and HBcAg negative. Modes of transmission were congenital 56.4% as well as others including: through homodialysis, continuous dental services, tattooing in addition to suspected intercourse all together comprised about 43.6%.
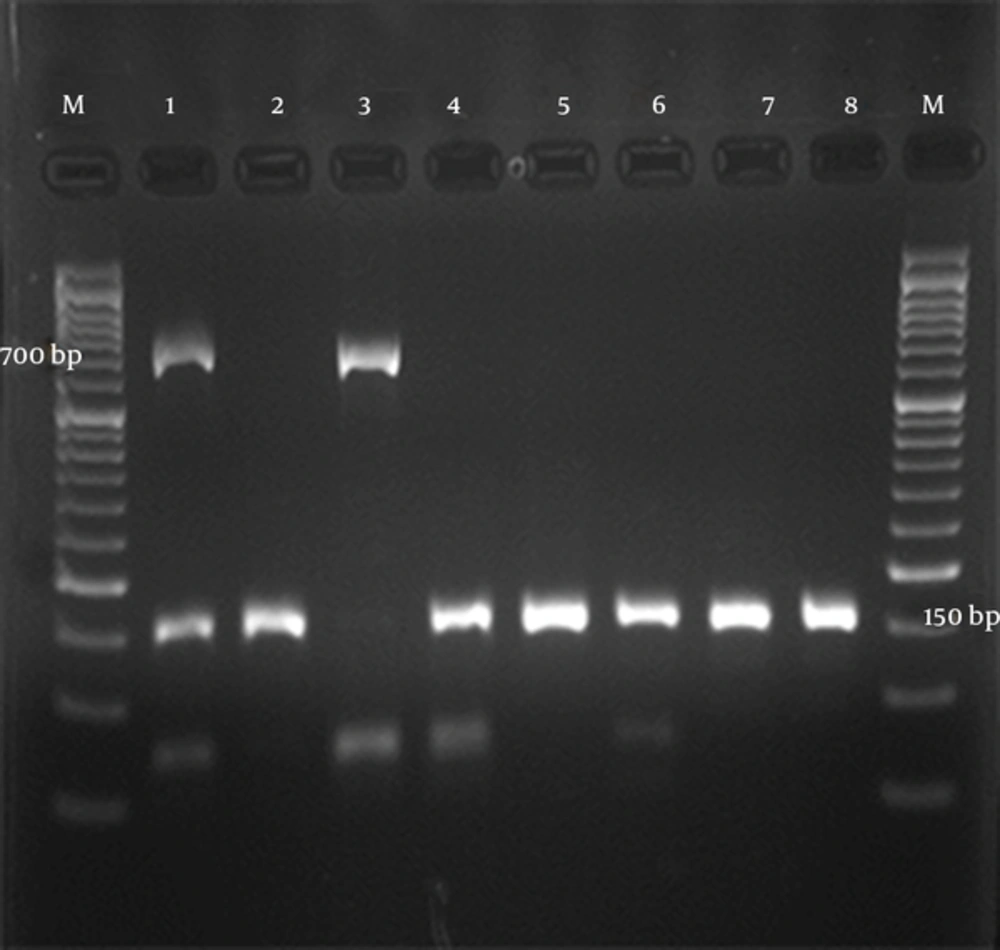
All sera samples contained HBV DNA which were extracted by above mentioned protocol and were then subjected onto multiplex PCR. Findings were evident for genotype D in 154 specimens (94.5%) and 9 (5.5%) had mixture of C/D genotype (Figure 1).
5. Discussion
In this study, similar to the other researches carried out in different parts of Iran and Middle East, genotype D was predominant. According to the data published roughly 1% - 3% of Iranian population are assumed to be carriers of HBV but with various frequencies in different provinces for instance; in Fars province this figure is 1.7% while for Sistan and Baluchestan is more than 5% [19]. Eight genotypes of HBV (A - H) are differentiated according to at least 8% difference in complete nucleotide sequences or more than 4% in S gene [20]. Genotypes of HBV do not indicate any particular geographic distribution. The latter pattern has shifted greatly specially in countries where more immigration takes place and it is anticipated that with escalation of immigration worldwide the former geographic distribution may seem more fluctuating. Hence, there are possibilities of recombination between genotypes e.g. A with D or A with C or B with C and so on. Although, there are certain structural and clinical differences between genotypes but recombination with other strains would create more variations and definite evidences support the concluding theory. Nevertheless in most of the studies, in spite of geographical pattern of genotypes distribution, regretfully only genotypes A with D and B with C have been compared and therefore, there is no clear consensus regarding virulence of different genotypes. Ideally, we need a prospective study in which neonates who have congenital infections with various genotype of A, B, C, D, E, F and G be taken under scrutiny up to youth in order to answer questions on differences between genotypes after chronic infection of HBV [21].
In regions where HBV is endemic, infection with more than one genotype frequently ends up to recombinant strain and possibility of co-infection or super-infection with other genotypes in a particular host. Furthermore, mixture of different genotypes in patients suffering from chronic hepatitis B (CHB), in comparison with those who are infected with a single genotype, is related to higher viral load and acceleration of HBV replication in vitro. In a study by Cao in China, the prevalence of genotype mixture in HBsAg carriers who were asymptomatic, hepatocellular carcinoma (HCC) sufferers and CHB victims was reported as: 5.4%, 10.6% and 13.7% respectively. Genotype mixture (mostly B and C) in comparison with C alone is correlated with more viral load and a more severe disease [5]. In addition according to above report co-infection and super-infection with various genotypes has poorer prognosis of HBV. In another study by Fang et al., a mixture of genotypes C/D in China has been documented [22]. Similarly, Noorali et al. in addition to genotype D which is predominant in Pakistan, have described co-infection of genotypes B/D as well as C/D [23].
As far as Iran is concerned, information on HBV genotype is limited nonetheless, Mohammadnejad et al. in eastern Azerbaijan [24], Moradi et al. in Golestan province [25], Eftekhari et al. in southern provinces [18], Sharifi et al. in Kerman, Esfahan and Yazd [26] as well as Dokanehiifard and Bidmeshkipour in Kermanshah province [27] have unambiguously announced genotype D as prevailing one. As discrepancy of genotypes in a certain country is foreseeable, so HBV genotyping sounds necessary in different regions.
As a matter of fact, at beginning classification of HBV was based on complete nucleotide sequencing of genome but later on the former gradually changed to sequencing of single genes or parts of them in order to facilitate the comparison between different strains. In search for alternate techniques in lieu of sequencing led to trial of Restriction Fragment Length Polymorphism (RFLP) for PCR products of HBV. Therefore, S gene or combination of both pre S and S genes are employed for genotyping. In a study by Mizokami et al. who compared complete genome by RFLP analysis with that of S gene, concluded the latter alone is sufficient for differentiation between all six genotypes i.e. A - F [28]. Another procedure used for genotyping was analysis of PCR products i.e. hybridization after PCR or so called line probe assay. Naito et al. explicated genotyping via PCR by means of specific primers [29]. Similarly, Usuda et al. through utilization of genotypes dissimilarity in pre S2 gene, applied monoclonal antibodies against specific epitopes and for discrimination between genotypes A to F they used ELISA [30]. In another investigation, Alavian et al. extracted genotype D from 109 patients by another protocol known as: INN-LiPA hybridation [19]. However, Mohammadnejad et al., in eastern Azerbaijan for HBV genotyping put into practice sequencing followed by analysis of products [24]. In contrast to former techniques, Dokanehiifard and Bidmeshkipour in Kermanshah employed RFLP for genotyping [27] whereas, Moradi et al. by utilization of specific commencers of A-F genotypes could differentiate between strains in samples obtained in Golestan province [25].
As far as multiplex PCR is concerned, it is useful for rapid diagnosis plus deletion of repeating gene in a long chain, so in this procedure couple of primers is added to a PCR cocktail aiming in finding of different genes that leads to PCR products with various sizes. Multiplex PCR is advantageous in terms of saving time and reagents as well. In present study we employed multiplex PCR due to its sensitivity, specificity and cost effectiveness besides this protocol was approved by Kirschberg et al. in their study [9]. Therefore, we extracted DNA from sera of HBV infected individuals and then through PCR, differentiation of A-F genotypes by a single reaction was obtainable.
Furthermore, this technique is quite appropriate for epidemiologic as well as diagnostic purposes in areas like Iran with relatively high frequency of HBV. Multiplex PCR is suitable for diagnoses of co-infections with various genotypes as well. The current study can also lead to find new ways for remedy of HBV victims and further investigations would aid in identification of various sera and subtypes of the virus in question. Moreover, such investigation on HBV serotyping would assist in reconstruction of history of evolution of certain strains and of course would complement our knowledge about genetic information of HBV.