1. Background
Few retroviruses are associated with human tumors (1). Human T-cell lymphotropic virus (HTLV) type 1 causes some types of lymphoma/leukemia T cells in adults (ALT), as well as a shambling disease of the nervous system called tropical spasmodic paralysis (TSP). Studies proved the association between this virus and a wide range of rheumatologic diseases including uveitis, polymyositis, thyroiditis, autoimmune, pneumonia, and arthritis. Moreover, a number of fatal and opportunistic infections in patients with T-cell leukemia are diagnosed (2, 3). Although these illnesses are not very common, they are usually serious and cause progressive disabilities or death (4). Therefore, infection with this virus is considered as a major concern for public health (5).
Patients with thalassemia need infusion of four to six blood units per month, and therefore, the risk of blood-borne diseases are caused by viruses such as HTLV-1 (6).
Although the exact number of people with HTLV-1, 2 is not known, it is estimated that around 15 to 20 million people are infected with the virus worldwide (7). The endemic regions of the world for a high prevalence of HTLV-1 include North-East Japan, several sub-Saharan African countries, Central and South America, certain regions of Iran, Melanesia, Taiwan, and the United States (8).
In Iran, this virus is found in isolated pockets in which HTLV-Ι infection is endemic (North Khorasan, the Northeastern province of Iran) (9).
In a study conducted by the blood transfusion organization of Iran on 82,689 blood donors in 27 provinces, the Western blot testing was positive in 0.085% of donors. In the current study, the highest prevalence of HTLV was observed in Fars, Tehran, and Qom provinces with 33%, 31%, and 30%, respectively. According to the current study, the prevalence of HTLV-1 infection in some provinces of Iran was significant; therefore, screening test on blood donors are necessary (5).
The previous reports show that HTLV-I is endemic in the Northeastern Iran, particularly in Mashhad, Sabzevar, and Neyshabour cities. The prevalence of HTLV-I infection in Mashhad, Neyshabour, and Sabzevar is 2.1%, 3%, and 1.6%, respectively (2, 3). However, the virus is less frequent in other parts of Iran including Urmia (0.34%), the Northwestern, and Chaharmahal-Bakhtiari province (0.62%) in the Southwestern Iran (1-5, 7, 8, 10, 11).
The HTLV-1 virus is transmitted through lactation feeding, sexual contact, blood transfusion, and the sharing infected syringes. Cellular blood products are the main source of transmission of the virus (11). Of these, the recipients of repeated blood transfusions (i e, thalassemia, hemophilia, hemodialysis) are at high risk of infection (12, 13). It should be noted that there is no effective treatment for the related diseases, except uveitis (low probability), the reduction of morbidity and mortality associated with the virus relies on successful prevention (6).
The routine diagnosis of HTLV infections is based on serological techniques such as the enzyme-linked immunosorbent assay (ELISA) and Western blot. However, among the samples infected with HTLV-1, 2, the ratio of uncertain results is high (14). The ELISA and Western blot methods show inaccurate serum results and cannot detect the early infection when the immune response is undermined. Moreover, the normal immunological methods cannot detect acute infections of these viruses (14). TaqMan real-time polymerase chain reaction (RT-PCR) is a quick and specific technique to detect the quantity of target virus. In this method, the inlet fluorescence probes specifically allow the detection of proliferative products directly related to the number of target copies (15).
Therefore, based on the study by Jalaeikhoo et al. (16), PCR was more accurate to diagnose HTLV-1 infection compared with ELISA as a diagnostic method.
2. Objectives
The current study aimed at detecting proviral HTLV-1 by TaqMan RT-PCR.
3. Methods
3.1. Sample Collection
In the current study, 80 blood samples from patients with thalassemia major referred to Tonekabon Shahid Rajaee Hospital from September to November 2015 were collected for blood counting (CBC) and blood transfusion. Of these 80 subjects, 47% were male and 53% female. Majority of them were 20 - 30 years old. Blood samples were taken daily in the Hospital and transferred to the university lab at -18°C and stored at -20°C.
3.2. DNA Extraction
Molecular biological system transfer kit (MBST) purchased from Cinna Gen company, Iran was used to extract the genomic DNA from blood cells. This kit contains four types of binding buffer, wash buffer, elution buffer, lysis buffer, and proteinase K enzyme. All buffers and columns should be kept at room temperature. Proteinase K was dry in this kit and kept at 4°C.
3.3. Qualitative and Quantitative Analysis of DNA
Due to the importance of the DNA extraction accuracy, all of the extracted DNAs were molecularly screened and then, quantitatively analyzed.
After extraction of DNA per day, according to the number of extracted samples, 7% - 21% agarose gel cements were prepared for the qualitatively analysis of DNAs. Each sample was loaded with buffer lining inside the well and electrophoresis was performed for all specimens.
In order to quantitatively inspect the cuvette, 1 μL of the sample, along with 49 μL of distilled water were inserted into a cuvette, and placed inside the biophotometer® (Eppendorf-Germany). Optical absorbance of the samples was read at 260 nm for sample concentration. Additionally, to determine sample purity, 260/280 and 260/230 wavelengths were also investigated. The remaining samples were kept at -20°C.
3.4. Positive and Negative Control
Positive and negative controls were used to obtain information about the efficiency and precision of the reaction. The current study was conducted to determine the mixture performance of the reaction and primers. The positive control sample used in the current study, due to the inaccessibility of the HTLV-1 specimen, was produced by artificial creation of the tax gene. The HTLV-1 tax gene sequence with accession: AY040796.1 was taken from the NCBI (the national center for biotechnology information) website and ordered to be manufactured by Nedaye Faan company. In order to determine the absence of contamination, a negative control without DNA was used; instead, buffer was added.
3.5. TaqMan RT-PCR Test
The RT-PCR TaqMan reaction was performed according to a study previously performed by Moens et al. (14) and the sequences of probes and primers used in the current study were planned according to the above study. These primers and probes are dedicated to the HTLV-1 protected tax area (Table 1).
| HTLV-1 Primers | DNA Sequencing (5’ → 3’) |
|---|---|
| Forward: (HTLV1-IF) | (5’- GGGTGGGTTCCATGTATCCA -3’) |
| Reverse: (HTLV1-IR) | (5’- CTCCTTCCCCACCCAGAGATT -3’) |
The 6- carboxy fluorescein- AGGAGGGTGGAATGTTGGGGGTTGTATG-BHQ1 was used to obtain duplicate signals from fluorogenic probes biconvex.
First, in a 1.5 mL microtube 12.5 µL of 2XTaqMan universal PCR master mix, 1 µL of HTLV-1 primer, and 0.5 µL of HTLV-1 probe with a buffer were mixed according to the number of samples. Since the total volume of the mixture should be 25 μg in each 0.2-microtube, 20 μL of the mixture was added to the microtube. Then, 51 μg DNA, extracted from each sample, and 6 µL dH2O were added to the corresponding microtube.
Then, sterilized 0.2-mL microtubes were provided based on the number of samples and placed in the specific block of the device and numbered, and then the instructions were given to the device.
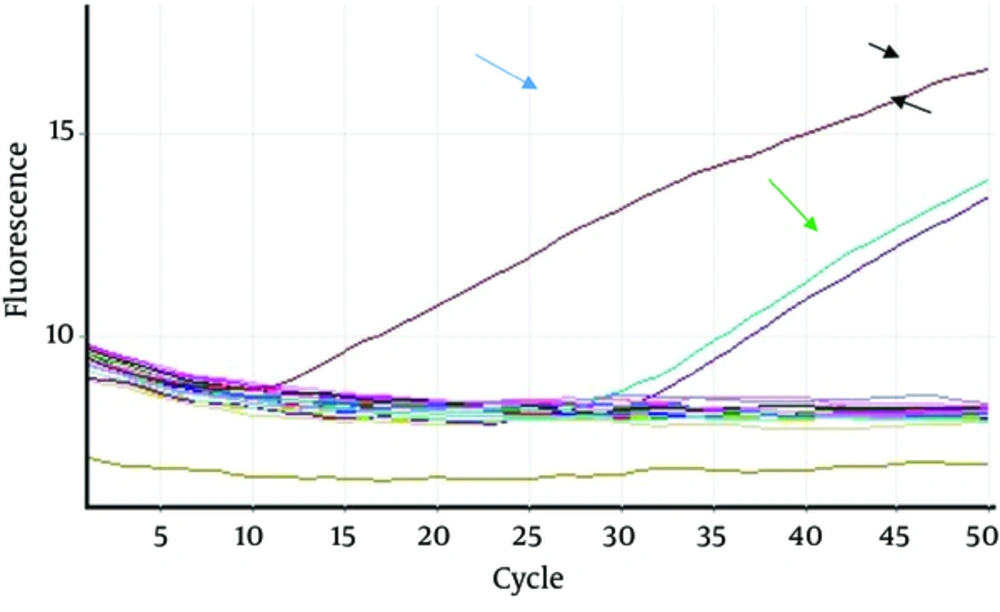
At this point, the response to data replication and analysis in the green channel was performed by the Corbett 6000 machine. The temperature pattern of the duplicate reaction was 95°C for 10 minutes for the initial activity step, then 50 cycles at 95°C for 15 seconds, and 60°C for one minute. After completing the task, the results were interpreted based on the obtained peaks.
4. Results
4.1. DNA Analysis Results
Due to the confirmation of DNA extraction in terms of presence and health, all extracted DNAs were analyzed qualitatively and its accuracy is visible in the below image and a DNA strip was observed without any smears.
4.2. DNA Quantitative Analysis Results
The average absorption of spectrophotometer for nucleic acid extracts was as follows:
As it was observed, the optical absorption ratio at 260/280 nm was 1.85, indicating that the sample was protein-free, and at 260/230 nm, it was 1. 57. In this case, almost no carbohydrates existed in the DNA sample.
4.3. Results of the TaqMan RT-PCR
The study population included 80 patients with β-thalassemia major and all of them lived in Tonekabon, where they referred to Shaheed Hospital at least once a month to receive blood. As shown in Figures 1 and 2, eighty of the genomic DNAs were extracted from blood and two (2.5%) of them were positive for TaqMan RT-PCR to detect the virus genome, of which two were female, 22 and 26 years old. Both were single. The remaining 78 samples (97.5%) were negative. Due to the low number of positive samples, statistical analysis was not performed.
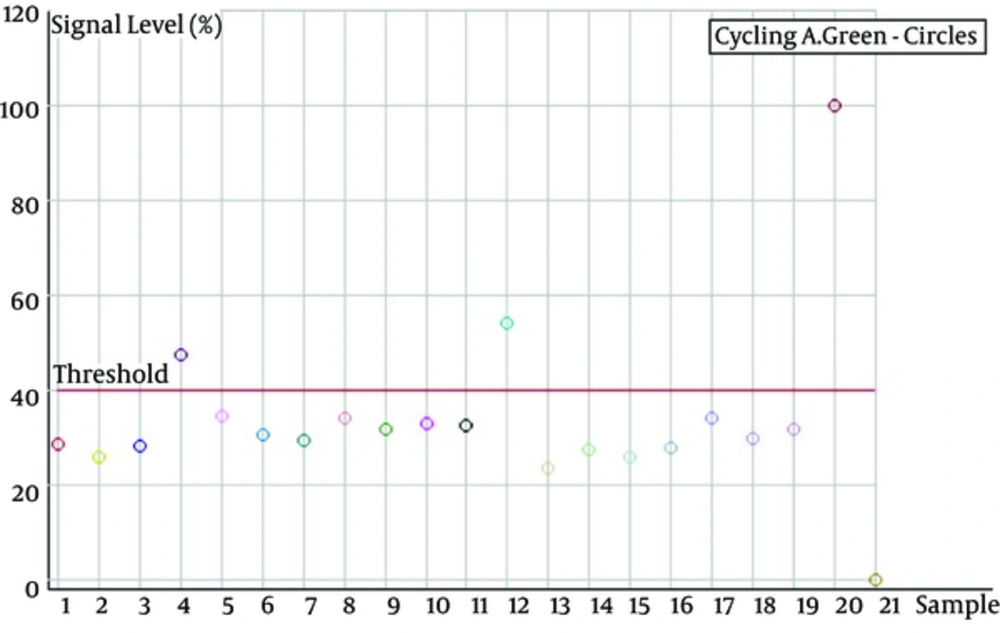
The production level diagram of the TaqMan RT-PCR signal; asshown in the above diagram, the sample 20 is lower than other samples and it is related to the negative control; and sample 20, which is higher than the other samples is positive control , The samples of 4 and 12 samples are not HTLV-1-infected and other samples are infected with HTLV-1.
5. Discussion
HTLV-1 virus infects T-cells and is easily transmitted through blood and blood products such as whole blood and platelets. Since HTLV-1 infection is chronic and untreatable, standard diagnostic and prevention, treatment, and adequate care should be provided. The best prevention method is to avoid using blood containing positive HTLV-1 (11, 12).
In the current study, two (2.5%) subjects were positive out of 80 patients by TaqMan RT-PCR assay similar to the study by Moradi et al. (12), in Gorgan that out of 181 patients with thalassemia, 28 subjects (14.9%) were positive with ELISA, while only 4.4% of them were approved by Western blot testing. Karimi et al. (11), compared the prevalence of serum infection of HTLV-1 between two high risk (thalassemia and hemodialysis patients) and healthy groups using ELISA and Western blot in Chaharmahal-Bakhtiari province from 2005 to 2006.The Western blot test results showed that 24 of 27 (89%) ELISA-positive samples from 800 cases (9).
Another study in Mashhad on 100 serums reported that the prevalence of HTLV1 was 2% in patients undergoing hemodialysis and blood donors (3). In the systematic review and meta-analysis by Hedayati-Moghaddam and Amini on Iranian patients with HTLV1, result showed that HTLV-1 infection prevalence among patients with thalassemia was 4.1% (13). In Another study in Mazandaran province, Iran on 288 serum samples collected from patients with thalassemia, the prevalence of HTLV-Ι in the patients was 1.4% (6). A cross sectional study by Jalaeikhoo et al. (16), in the hematology and medical oncology department at army Hospital on 211 serums in January 2015 showed only two positive cases (0.9%).
Trevino et al. (17), investigated the prevalence of HTLV-1 and -2 infection in 6460 foreign patients admitted to 16 Hospitals in Spain. In their study, only nine patients had HTLV antibodies by ELISA. Of the nine positive cases, four were HTLV-1 and five HTLV-2 using Western blot technique. The prevalence of HTLV-1 and -2 infections was 18% in Southern Chile in patients with malignant hematological diseases of which 27% had chronic lymphoproliferative disorders, as Barrientos et al. (18), reported. Monavari et al. (10), reported that the prevalence of HTLV-1 among patients with malignant hematological diseases in Tehran, Iran was 12%.
The large difference between the results of ELISA (the main test to diagnose this infection in most parts of the world including Iran) and those of Western blot as well as the existence of unspecified test profiles of Western blot in some studies reveal the need for other diagnostic methods based on molecular techniques. TaqMan RT-PCR is a fast and proprietary technique to detect target viral infections. In this method, the fluorescent probes create a specific input that allows the recognition of reproductive products directly related to the number of target copies (15, 19, 20).
In a study by Li and Green (21), using a pair of oligonucleotide primers and special probes, a quantitative determination of the two types of HTLV-1 and -2 mRNAs was performed using RT-PCR. Andrade et al. (22), in Brazil used singleplex RT-PCR as a confirmatory test to detect HTLV-1 and -2 infections in blood donors. In fact, the results of their study demonstrated higher sensitivity and specificity of the applied molecular testing, especially in the diagnosis of HTLV-1, 2 infections. In a study by Besson and Kazanji (15), for the first time, RT-PCR multiplex was used based on the Beacon probes to differentiate and simultaneously determine the HTLV-1, 2, 3 viruses. They concluded that this method could be used for epidemiological studies in Africa and other countries where HTLVs are endemic.
According to many observations, HTLV-1 uses a non-toxic strategy to resist activated immune system. The virus is propagated in the form of the provirus, stimulating cell division, and replicating the colony of the infected cells by the virus (22). HTLV-1 is unusually low in genetic variation among its isolates, since replication of HTLV-1 in HIV-infected cells rarely occurs by reverse transcriptase enzyme and often occurs through cell DNA polymerase. Furthermore, the progression of ATL disease is essentially related to provirus loading. These findings suggest that proviratory loading plays an important role in the pathogenicity of the HTLV-1 virus (22).
The current study aimed at detecting and determining the propulsion load of HTLV-1 genome based on the intrigued tax gene in the genome of lymphocytes in patients with β-thalassemia major. The TaqMan RT-PCR method showed that in the current study, out of 80, two cases (% 2.5) of the patients were infected with HTLV-1.
5.1. Conclusions
Due to sensitivity, simplicity, higher speed, and the ability to simultaneously detect HTLV and lower fluorescent-based molecular methods than other molecular and serological methods, TaqManReal-Time PCR technique can be used to diagnose HTLV-1 infection in blood banks.
-thumbnail.webp)