1. Introduction
Acinetobacter baumannii is a Gram-negative coccobacillus bacteria, aerobic and nonfermenting that belonging to the Moraxellaceae family these Species are opportunist pathogens with increasing relevance in both community-acquired and hospital-acquired, especially among patients in intensive care units and risk factor patient [1, 2]. A. baumannii have been implicated in different nosocomial infections, including ventilator-associated pneumonia, endocarditis, meningitis, wounds, blood and infections of the skin, soft tissues, urinary tract infections, and those originating from other hospital environment [3]. A. baumannii can be isolated from numerous sources such as soil, water, animals, and humans, where ever presented in health care institutions or nosocomial infections or even in environmental surfaces, it will be difficult to control it [4, 5].
Antimicrobial resistance owing from enzymatic degradation, modification of targets, or bacterium in hospital environments are responsible for epidemics of A. baumannii in hospital-acquired species [6]. Current study show resistance to nearly all major classes of antibiotics, especially including broad-spectrum penicillin’s, cephalosporin’s, carbapenems, aminoglycosides, fluoroquinolones, chloramphenicol, and tetracycline’s. Today there are global distributions of multidrug resistant (MDR) in clinical isolates all over the word [7].
Increasing of carbapenem-resistance and β-lactamases resistance in A. baumannii tend us to study genotypic and phenotypic resistance pattern in detected specimens which isolated from patients hospitalized in different sections of hospital [8, 9].
This study helps us to clarify different mode of A. baumannii antibiotic resistance pattern and its epidemiology in different hospitals. The current review was conducted to elucidate of antibiotic resistance in A. baumannii that detected from hospitalized patients in Isfahan Iran.
2. Methods
2.1. Bacterial Isolates
A total of 150 isolates of A. baumannii was collected from clinical specimens from 3 major hospitals in Isfahan, Iran. Finally 150 clinical samples from individuals suffering from different types of infections were selected.
These isolates were characterized and confirmed in the laboratories of the corresponding hospitals through routine microbiological and biochemical tests such as IMVIC, urease, TSI, OF, MRVP, SIM, catalase, oxidase, and growth at 37°C and 42°C. The confirmed samples were stored at 30% glycerol at -70°C.
A. baumannii including blood (n = 38), wound (n = 33), tracheal secretion (n=10), meningitis (n=6) and urinary infection (n = 51) which were collected from patients hospitalized in ICU, orthopedic and internal section. All samples were directly cultured in to 7% sheep blood agar (Merck, Darmstadt, Germany) and incubated aerobically at 37°C for 48 hours. After incubation, suspicious colonies were examined by using laboratory techniques appropriate for diagnosing A. baumannii species [10-12].
2.2. Antibiotic Susceptibility Testing
A. baumannii isolates was selected and the antibiotic resistance pattern was performed by a disk diffusion method on Mueller-Hinton agar.
A. baumannii isolates were tested for sensitivity to tetracycline (30 µg), ceftazidime (30 µg), ciprofloxacin (30 µg), cotrimoxazole (25 µg), tobramycin (10 µg), Chloramphenicol (30 µg), Norfloxacin (10 µg), amikacin (30 µg), gentamycin (10 µg), Rifampin (5 µg), cefalotin (30 µg), Streptomycin (10 µg), Trimethoprim (5 µg), Levofloxacin (5 µg), Imipenem (10 µg), meropenem (10 µg), Nitrofurantoin (300 µg), azithromycin (15 µg), erythromycin (15 µg), by the Kirby-Bauer disk diffusion method. (MAST, Merseyside, England), according to clinical and laboratory standards institute (CLSI) 2011. A. baumannii ATCC 19606 was used as the positive control strain [13-15].
2.3. Molecular Typing and DNA Extraction
Detection of the main groups of genes which are effective in A. baumannii resistance were performed by using M-PCR. The primers used are shown in Table 1. DNA extraction was carried out using the phenol chloroform method.
| Primers | Nucleotide Sequences (5’ - 3’) | Expected Amplicon Size, bp |
|---|---|---|
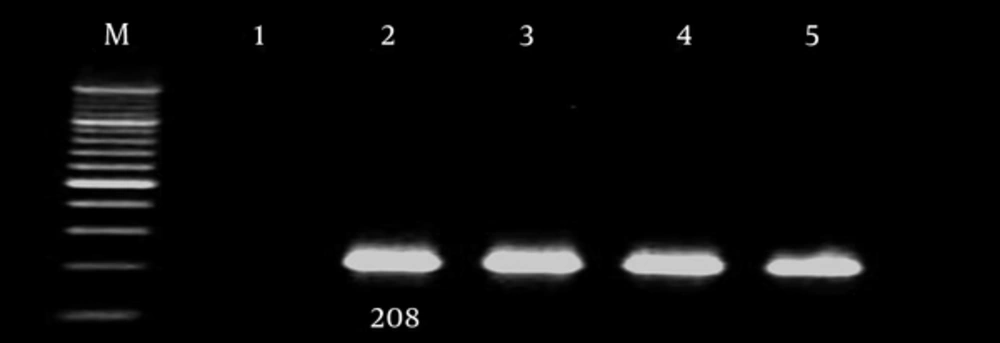
| 16S-23S ribosomal DNA | F- CATTATCACGGTAATTAGTG | 208 |
| R- AGAGCACTGTGCACTTAAG | ||
| aadA1 | (F) TATCCAGCTAAGCGCGAACT | 447 |
| (R) ATTTGCCGACTACCTTGGTC | ||
| aac(3)-IV | (F) CTTCAGGATGGCAAGTTGGT | 286 |
| (R) TCATCTCGTTCTCCGCTCAT | ||
| sul1 | (F) TTCGGCATTCTGAATCTCAC | 822 |
| (R) ATGATCTAACCCTCGGTCTC | ||
| blaSHV | (F) TCGCCTGTGTATTATCTCCC | 768 |
| (R) CGCAGATAAATCACCACAATG | ||
| CITM | (F) TGGCCAGAACTGACAGGCAAA | 462 |
| (R) TTTCTCCTGAACGTGGCTGGC | ||
| cat1 | (F) AGTTGCTCAATGTACCTATAACC | 547 |
| (R) TTGTAATTCATTAAGCATTCTGCC | ||
| cmlA | (F) CCGCCACGGTGTTGTTGTTATC | 698 |
| (R) CACCTTGCCTGCCCATCATTAG | ||
| tet(A) | (F) GGTTCACTCGAACGACGTCA | 577 |
| (R) CTGTCCGACAAGTTGCATGA | ||
| tet(B) | (F) CCTCAGCTTCTCAACGCGTG | 634 |
| (R) GCACCTTGCTGATGACTCTT | ||
| dfrA1 | (F) GGAGTGCCAAAGGTGAACAGC | 367 |
| (R) GAGGCGAAGTCTTGGGTAAAAAC | ||
| Qnr | (F) GGGTATGGATATTATTGATAAAG | 670 |
| (R) CTAATCCGGCAGCACTATTTA | ||
| Imp | (F) GAATAGAATGGTTAACTCTC | 188 |
| (R) CCAAACCACTAGGTTATC | ||
| Vim | (F) GTTTGGTCGCATATCGCAAC | 382 |
| (R) AATGCGCAGCACCAGGATAG | ||
| Sim | (F) GTACAAGGGATTCGGCATCG | 569 |
| (R) GTACAAGGGATTCGGCATCG | ||
| Oxa-51-like | (F) TAATGCTTTGATCGGCCTTG | 353 |
| (R) TGGATTGCACTTCATCTTGG | ||
| Oxa-23-like | (F) GATCGGATTGGAGAACCAGA | 501 |
| (R) ATTTCTGACCGCATTTCCAT | ||
| Oxa-24-like | (F) GGTTAGTTGGCCCCCTTAAA | 246 |
| (R) AGTTGAGCGAAAAGGGGATT | ||
| Oxa-58-like | (F) AAGTATTGGGGCTTGTGCTG | 599 |
| (R) CCCCTCTGCGCTCTACATAC | ||
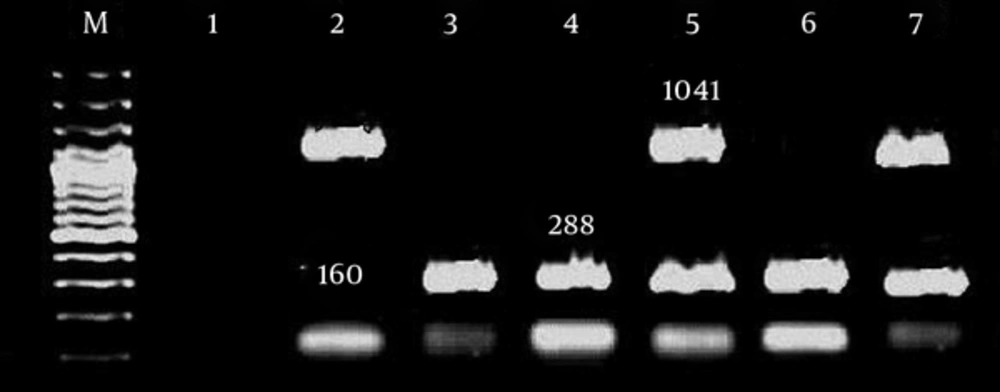
| IntI | F: CAG TGG ACA TAA GCC TGT TC | 160 |
| R: CCC GAC GCA TAG ACT GTA | ||
| IntII | F: TTG CGA GTA TCC ATA ACC TG | 288 |
| R: TTA CCT GCA CTG GAT TAA GC | ||
| IntIII | F: GCC TCC GGC AGC GAC TTT CAG | 1041 |
| R: ACG GAT CTG CCA AAC CTG ACT |
Primer pairs shown in Table 1 were used to detect antibiotic resistance genes, including aadA1 (streptomycin resistance), aac(3)-IV (gentamycin resistance), sul1 (sulfonamide resistance), blaSHV and CITM (beta-lactam resistance), cat1 and cmlA (chloramphenicol resistance), tet(A) and tet(B) (tetracycline resistance), dfrA1 (trimethoprim resistance), qnr (quinolone resistance), imp, vim, and sim (carbenicillin resistance), and Oxa-23-like, Oxa- 24-like, Oxa-51-like, and Oxa-58-like (oxacillin resistance). Genomic DNAs of A. baumannii isolates were extracted using Genomic DNA Purification Kit (fermentas Lithuania) and implemented as a template DNA in PCR amplification of the intended fragments. The PCR programs were set according to the size of the fragments as follows:
A multiplex PCR containing 50 μL of ingredients (5 μL of 10X PCR buffer, 2 mmol of MgCl2, 150 μmol of dNTPs mix, 0.5 μmol of F and R primer pairs (for each gene), 1.5 unit of Taq DNA polymerase, and 2 μL of each DNA sample) was used to detect aadA1, aac (3)-IV, sul1, blaSHV, CITM, cat1, cmlA, tet (a), tet (B), dfrA1, and qnr. The thermal program was a cycle of 94°C for 6 minutes, 33 repetitive cycles of 95°C for 70 seconds, 55°C for 65 seconds, and 72°C for 90 seconds, and a final cycle of 72°C for 8 minutes [15-18]. Strains of E. coli O157:K88ac:H19, CAPM 5933 and E. coli O159:H20, CAPM 6006 were used as the positive controls. The amplification of imp, vim, and sim genes was carried out in a multiplex PCR in a volume of 50 μL containing 5 μL of 10X PCR buffer, 1.5 mmol of MgCl2, 100 μmol of dNTPs mix, 1.0 μmol of forward and reveres primer pairs (for each gene), 1.0 unit of Taq DNA polymerase, and 2.5 μL of each DNA. The program was set to a cycle of 95°C for 4 minutes, 30 repetitive cycles of 94°C for 45 seconds, 58°C for 60 seconds, and 72°C for 40 seconds, and a final cycle of 72°C for 5 minutes [19]. Oxacillin resistance genes (Oxa-23-like, Oxa-24-like, Oxa-51-like, Oxa-58-like) were detected in a 50 μL multiplex PCR with 5 μL of 10X PCR buffer, 2.5 mmol of MgCl2, 200 μmol of dNTPs mix, 0.5 μmol of forward and reveres primer pairs (for each gene), 1.5 unit of Taq DNA polymerase, and 2.0 μL of each DNA of each isolate. The program included a cycle of 94°C for 5 minutes, 32 repetitive cycles of 95°C for 50 seconds, 60 °C for 60 seconds, and 72°C for 70 seconds, as well as a final cycle of 72°C for 10 minutes [16-20].
50 microliters of M-PCR products were resolved on a 1.5% agarose gel containing 0.5 mg/mL of ethidium bromide in Tris-borate-EDTA buffer at 90 V for 1 hour. Finally the products were examined with ultraviolet illumination [20].
2.4. Analyzing the Presence of Integrons
Classes 1, 2, and 3 integrons were detected using the primers shown in Table 1 in the multiplex PCR. The reaction volume was set to 25 μL containing 2.5 μL of 10X PCR buffer, 1.5 mmol of MgCl2, 200 μmol of dNTPs mix, 0.5 μmol of forward and reveres primer pairs, 1.0 unit of Taq DNA polymerase, and 2.5 μL of each DNA sample. Thermal program was set to a cycle of 94°C for 6 minutes, 35 repetitive cycles of 94°C for 1 minutes, 56°C for 1 minutes, and 72°C for 45 seconds, as well as a final cycle of 72°C for 6 minutes. Since the positive control sample was not used in regard with detection of some genes, to confirm or reject the PCR results, the PCR products of the primary positive samples were purified using a PCR product purification kit (Roche Applied Science, Germany) and sent to the Macrogen Co. (South Korea) for sequencing. The PCR amplifications were carried out in an Eppendorf, Mastercycler® 5330 (Eppendorf-Netheler-Hinz GmbH, Hamburg Germany). The amplified fragments were visualized on ethidium bromide containing 1.5% of agarose gels, in which 15 μL of each product was loaded, in addition to 100 bp DNA marker (fermentas-Lithuania), on a UV transluminator (UV itech, England) after electrophoresis at 90 V for approximately 45 minutes.
24-like, Oxa-51-like, and Oxa-58-like (oxacillin resistance). Genomic DNAs of A. baumannii isolates were extracted using Genomic DNA Purification Kit (fermentas Lithuania) and implemented as a template DNA in PCR amplification of the intended fragments. The PCR programs were set according to the size of the fragments as follows:
A multiplex PCR containing 50 μL of ingredients (5 μL of 10X PCR buffer, 2 mmol of MgCl2, 150 μmol of dNTPs mix, 0.5 μmol of F and R primer pairs (for each gene), 1.5 unit of Taq DNA polymerase, and 2 μL of each DNA sample) was used to detect aadA1, aac (3)-IV, sul1, blaSHV, CITM, cat1, cmlA, tet (a), tet (B), dfrA1, and qnr. The thermal program was a cycle of 94°C for 6 minutes, 33 repetitive cycles of 95°C for 70 seconds, 55°C for 65 seconds, and 72°C for 90 seconds, and a final cycle of 72°C for 8 minutes [15-18]. Strains of E. coli O157:K88ac:H19, CAPM 5933 and E. coli O159:H20, CAPM 6006 were used as the positive controls. The amplification of imp, vim, and sim genes was carried out in a multiplex PCR in a volume of 50 μL containing 5 μL of 10X PCR buffer, 1.5 mmol of MgCl2, 100 μmol of dNTPs mix, 1.0 μmol of forward and reveres primer pairs (for each gene), 1.0 unit of Taq DNA polymerase, and 2.5 μL of each DNA. The program was set to a cycle of 95°C for 4 minutes, 30 repetitive cycles of 94°C for 45 seconds, 58°C for 60 seconds, and 72°C for 40 seconds, and a final cycle of 72°C for 5 minutes [19]. Oxacillin resistance genes (Oxa-23-like, Oxa-24-like, Oxa-51-like, Oxa-58-like) were detected in a 50 μL multiplex PCR with 5 μL of 10X PCR buffer, 2.5 mmol of MgCl2, 200 μmol of dNTPs mix, 0.5 μmol of forward and reveres primer pairs (for each gene), 1.5 unit of ribosomal DNA) through simple disc diffusion method. All the studied isolates demonstrated the multidrug resistance (MDR).
2.5. Statistical Analysis
The collected data were analyzed by SPSS statistical software (ver.16) using Chi-square and Fischer’s exact statistical tests at the confidence level of 95 percent.
3. Results
During this study, 220 samples of Acinetobacter were collected from different patients in internal section, orthopedic and ICU. One hundred and fifty samples of isolate were identified as A. baumannii (68.8%), 50 samples were Acinetobacter Lwoffii (27.7%) and 20 samples (9.09%) were identified as other Acinetobacter species. Among of isolated A. baumannii strains, all samples (100%) were multidrug- resistant. Results of this study showed that 79 samples of A. baumannii (52.6%) were resistant to three or more than three antibiotics and 97 samples (64.6%) showed resistance to more than two antibiotics. Also, none of resistant strains were showed complete resistance to all antibiotics. According to antibiotic resistance pattern, it was mentioned that, approximately 98.3% of samples were resistant to ciprofloxacin, 89.4% of isolates were resistance to ceftazidime and there were 87.3% resistance to tetracycline. Other antimicrobial resistance profiles of A. baumannii is shown in Table 2.
| Antibiotic | Sensitive Percent | Intermediate Percent | Resistance Percent |
|---|---|---|---|
| Tetracycline | 87.3 | 10.8 | 1.9 |
| Ceftazidime | 89.4 | 5.2 | 5.4 |
| Ciprofloxacin | 98.3 | 1.7 | 0 |
| Cotrimoxazole | 65 | 25.1 | 9.9 |
| Tobramycin | 53 | 35.4 | 11.6 |
| Chloramphenicol | 3.5 | 11.5 | 85 |
| Norfloxacin | 78 | 12.5 | 9.5 |
| Amikacin | 55.7 | 30.3 | 14 |
| Gentamycin | 79.5 | 18.3 | 2.2 |
| Rifampin | 45.8 | 40.8 | 13.4 |
| Cefalotin | 56.1 | 27.3 | 16.6 |
| Streptomycin | 34.3 | 7 | 58.7 |
| Trimethoprim | 73 | 13.6 | 13.4 |
| Levofloxacin | 55 | 20.5 | 24.5 |
| Imipenem | 43 | 38.8 | 18.2 |
| Meropenem | 66.5 | 29.4 | 4.1 |
| Nitrofurantoin | 3.2 | 13.5 | 83.3 |
| Azithromycin | 74 | 14.5 | 11.5 |
| Erythromycin | 65 | 17.9 | 17.1 |
Molecular typing of genes that related to A. baumannii antibiotic resistance pattern show the frequency of genes as dfrA1 (63.7%), sul1 (68.6%), aac(3)-IV (54.4%), tet(B) (22.4%), tet(A) (78.3%), aadA1 (15.4%), CITM (17. %), vim (12.2%), Qnr (17.1%), blaSHV (19.8%), sim (7.8%), Oxa-24-like (13.2%), Oxa-51-like (11.9%), Oxa-58-like (39.4%), Oxa-23-like (12.6%), imp (9.2%), cmlA (19%) and cat1 (8.6%) respectively. Frequency of integrons class I, II, III was (100%), (28%), (6.6%) respectively. There were a significant relationship between integron I gene and detected specimens. All of the specimens were contains integron class I. Frequency of other integron classes and its relationship with different groups of specimens are show in Table 3.
| Frequency Sample Type | Integron I | Integron II | Integron III |
|---|---|---|---|
| 38/Blood | 38 | 9 | 1 |
| 33/Wound | 33 | 10 | 2 |
| 51/UTI | 51 | 16 | 1 |
| 6/meningitis | 6 | 2 | 1 |
| 22/T. secretion | 22 | 5 | 5 |
| Total | 150 | 42 | 10 |
A. baumannii isolates which detected from different Patients in Prevalence of age show that the most frequent age range of the patients were between 61 - 70 age (57.9%) who were hospitalized in different section of hospitals. Also 69% of patients were women in this study.
In current study most of the integron class I and integron classes II were found among multidrug resistant A. baumannii strains. The results of our study were in concordance with similar studies around the world. Due to increased drug resistance, wider search and further study is necessary and can be useful to control hospital infections [21]. Gel electrophoresis of samples are show in Figures 1 - 3.
4. Discussion
Molecular typing of genes that related to A. baumannii antibiotic resistance pattern show the frequency of genes as dfrA1 (63.7%), sul1 (68.6%), aac(3)-IV (54.4%), tet(B) (22.4%), tet(A) (78.3%), aadA1 (15.4%), CITM (17. %), vim (12.2%), Qnr (17.1%), blaSHV (19.8%), sim (7.8%), Oxa-24-like (13.2%), Oxa-51-like (11.9%), Oxa-58-like (39.4%), Oxa-23-like (12.6%), imp (9.2%), cmlA (19%) and cat1 (8.6%) respectively. Frequency of integrons class I, II, III was (100%), (28%), (6.6%) respectively. There was a significant relationship between integron I gene and detected specimens. All of the specimens were contains integron class I. Frequency of other integron classes and its relationship with different groups of specimens were show in Table 3.
Different species of Acinetobacter have strong tendency to become resistant to antibiotics and are inherently resistant to some broad antibiotics, and on the other hand have a great ability to gain new mechanisms of resistance. Plasmids, transposons and integrons are important factors in the acquisition and transfer the mechanisms of antibiotic resistance [22].
Today, the spread of antibiotic resistance through integron structures with multiple antibiotic resistance has become a serious problem in treatment of infections caused by Acinetobacter baumannii. Due to the spread of antibiotic resistance in Acinetobacter baumannii, and since the city had not done a comprehensive study on antibiotic resistance of these bacteria, this study was conducted to show pattern of antibiotic resistance, genetic pattern of antibiotic resistance and tracking Integrons classes. All isolates examined in this study were MDR [23].
The highest rate of drug resistance related to tetracycline (87.3%), ciprofloxacin (98.3%), and ceftazidime (89.4). Similar studies have been conducted in different regions of Iran too. In a similar study conducted by Momtaz et al. (2015) in two large hospitals in Tehran, almost similar pattern was observed that 90.90 % of Acinetobacter baumannii isolated from human infection were resistance to tetracycline, 61.98% to trimethoprim, 51.23% were resistance to cotrimoxazole and between 9.91% to 31.40% of isolates were resistant to aminoglycoside compounds [24].
In a study done by Shakibaii on 50 isolates of A. baumannii which detected from the intensive care unit in Kerman hospitals, the antibiotic-resistant pattern show 73.3% resistance to imipenem, 66% to ciprofloxacin, 93.3% to piperacillin-tazobactam, 53.3% to amikacin, 93.3% to Cefepime and 100% reported to piperacillin [25].
In a study conducted by Aliakbarzade et al. (2014) in Tabriz hospitals, antibiotic resistance pattern of Acinetobacter baumannii isolated from patients were at a rate of 94% to kanamycin, 86% to gentamicin, 81% were resistant to amikacin and 63% were resistant to tobramycin. Different antibiotic resistance in various studies can be related to clinical sample types, or geographic region of the test [26].
Distribution genes encoding resistance to antibiotics was one of the goals of this study. The presence of this genes coding in antibiotic resistance was show in this 150 isolates. Molecular typing of genes that related to A. baumannii antibiotic resistance pattern show the frequency of genes as dfrA1 (63.7%), sul1 (68.6%), aac(3)-IV (54.4%), tet(B) (22.4%), tet(A) (78.3%), aadA1 (15.4%), CITM (17. %), vim (12.2%), Qnr (17.1%), blaSHV (19.8%), sim (7.8%), Oxa-24-like (13.2%), Oxa-51-like (11.9%), Oxa-58-like (39.4%), Oxa-23-like (12.6%), imp (9.2%), cmlA (19%) and cat1 (8.6%) respectively and integrons class I, II, III was (100%), (28%), (6.6%) respectively. Compared with this study, Huang et al. (2008) in China, reported distribution of β-lactamase genes that were evaluated in 21 isolates of A. baumannii. In this study all of the isolates contain blaOXA-51-Likeand just 8 strains contains blaOXA-23-Likegene [27].
Another study by Arezzo in Italy (2011) show that 93.8% of 114 isolates were resistance to imipenem and 71.1% of isolates contains blaOXA-23-Likegenes and 22.8% of isolates contains blaOXA-58-Like, Compared with our study they didn’t detect Oxa-24-like, Oxa-51-like, vim, sim and imp in their isolates [28].
Frequency of integrons class I, II, III in our study was (100%), (28%), (6.6%) respectively. There was a significant relationship between integron I gene and detected specimens. All of the specimens were contains integron class I.
Unlike these results in the study of Mirnejad et al. (2012) 82% of Acinetobacter baumannii isolated from hospitals in Tehran, carried Integrons Class II. There was a significant correlation between integrons and antibiotics resistance pattern to Cefepime, aztreonam, amikacin, ciprofloxacin, norfloxacin, ofloxacin and ceftazidime [29].
Also in a survey conducted by Mohammadi et al. presence of integrase gene I, II and III in XDR of Acinetobacter baumannii which detected from patients in Hamedan hospitals, was 97%, 31% and 0% respectively [30]. In a study in Taiwan, 72% of Acinetobacter baumannii just contains integrase gene class I [31].
In Koczura reviews (2014) in Poland only 63.5% of Acinetobacter baumannii contains integron class I. Similar to this study in Turkey (2014) only 33% of the isolates of the bacteria carry Integrons class I, and class II [32].
Because of the differences in the prevalence of class I integrons in two recent studies with this study and also similar studies in Iran, so it can probably relate to the verities of geographical area , the proper use and control of antibiotics in out of Iran [33].