1. Background
HTLV-1 is a human retrovirus and the etiological agent of multiple disorders [1]. Approximately, 10 to 20 million people are estimated to be infected by HTLV-1 worldwide. These individuals will eventually develop an often rapidly fatal leukemia, a debilitative myelopathy, or they may experience uveitis, infectious dermatitis, or other inflammatory disorders [2].
Screening HTLV-1 carriers is currently the main strategy for disease prevention [1]. Screening is generally performed through ELISA, a highly sensitive and specific method. HTLV-1 infection is confirmed by conducting Western blotting on specimens testing positive in ELISA [1, 3].
Gp46-I (162-214) is a linear peptide with 48 amino acids, which is derived from the conserved region of HTLV-1 gp46 envelope protein, and exhibits specific strong reactivity against HTLV-1 infected sera. It is non-reactive to HTLV-2 positive sera; however, it is used detect anti-HTLV-1 antibodies in ELISA and to differentiate between HTLV-1 and HTLV-2 infections in Western blotting [3-5]. Expression of peptides in E. coli is one of the cost-efficient methods for providing diagnostic reagents [6]. However, it is difficult to determine the expression of small peptides in E. coli Introducing a fusion tag may have a positive effect on peptide expression and purification [7]. Many of small peptides have been successfully expressed in E. coli using glutathione S-transferase (GST) tag as a fusion partner [8, 9]. In a previous study, we constructed a recombinant protein derived from both HTLV-I and -II, incorporating linear epitopes of gp46-I, gp46-II, gp21, and p19, which were connected together by a short flexible linker as a spacer arm between epitopes [10]. Generally, purification procedure requires dilution, dialysis steps, and concentration methods. Thus, in the present study, we aimed at proposing a method to reduce the purification time. We modified and synthesized the gene encoding HTLV-1 Gp46-I (162-214) according to E. coli codon usage without altering amino acid sequence. The synthetic DNA was subcloned into pGS21a vector and expressed as inclusion body purified and on-column refolded using Ni-NTA affinity chromatography. In addition, we assessed binding capacity of gp46-I(162-214) using ELISA.
2. Methods
This study was a descriptive research. All the required reagents were purchased from Sigma Chemical (Missouri, USA). They were of analytical grade unless otherwise indicated. The chimeric protein construction and nucleotide sequencing were performed using Biomatik (Wilmington, USA). Restriction enzymes were purchased from Takara (Kyoto, Japan). Plasmid DNA extraction and gel purification kits were obtained from iNtRON (Daejoen, Korea). T4 ligase, DNA marker, and isopropyl β-D-1-thiogalactopyranoside (IPTG) were purchased from Fermentase (Opelstr, Germany). Protein marker was purchased from SinaClon BioScience Co. (Tehran, Iran) and nickel-charged nitrilotriacetic acid (Ni-NTA) agarose resin from Qiagen (Valencia, USA).
2.1. Serum Samples
Serum samples were collected from individuals, who tested positive for HTLV-I (anti-HTLV-I positive) infection. All tests were performed using a commercial immunodiagnostic ELISA kit (HTLV-I/II ELISA 4.0, MP Biomedical, Santa Ana, California, USA). Sampling was conducted in Razavi Hospital, Mashhad, Iran. Control serum samples were also collected from healthy volunteers and tested with ELISA.
2.2. Construction of Expression Vectors
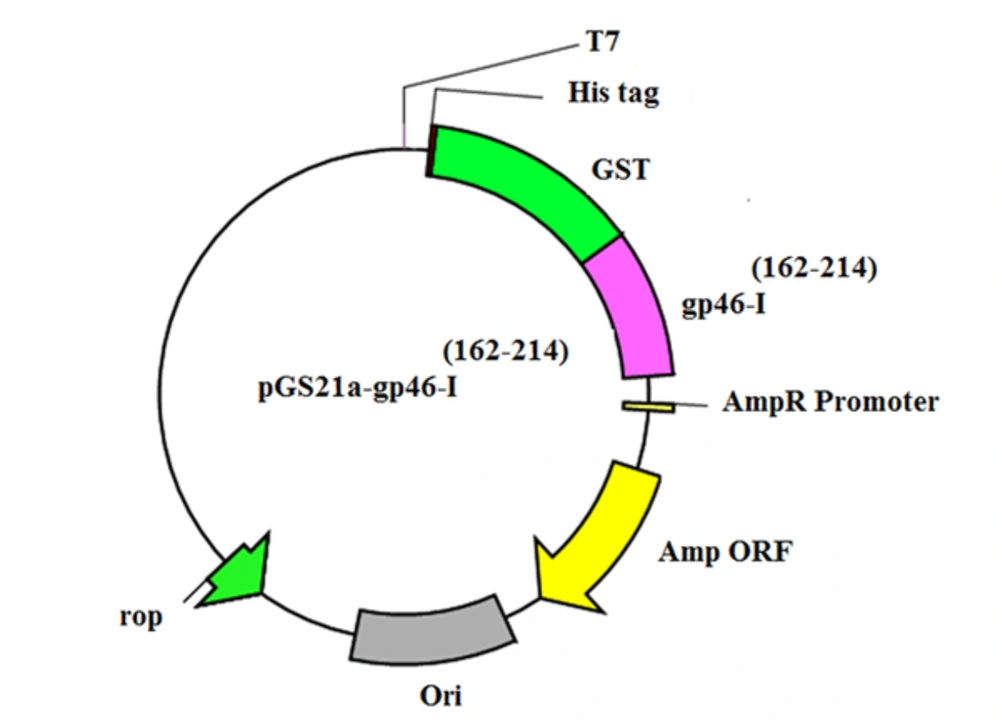
To create the expression vector for the gp46-I(162-214), the DNA encoding gp46-I(162-214) was optimized according to E. coli codon usage by JAVA codon optimization tool (http://www.jcat.de/).Rare codon analysis tools (https://www.genscript.com/tools/rare-codon-analysis) were used to analyze the codons; they were then synthesized by Biomatik (Wilmington, USA). The synthetic gene was sub-cloned into the BamHI/EcoRI sites of the pGS21a vector using standard protocols. The gp46-I(162-214) fused with a histidine tag (His-tag) and a GST tag; and enterokinase cleavage site at the N-terminus was expressed from the resulting plasmid, which is referred to as pGS21a-gp46-I(162-214).
2.3. Fusion Protein Expression
A single colony harboring pGS21a-gp46-I(162-214) plasmid was inoculated into 15 mL of Luria-Bertani (LB) medium with 100 µg/mL ampicillin and grown overnight at 37°C. The cultures were then diluted 1:100 into 50 mL of LB medium with 100 µg/mL ampicillin. Then the cells were constantly shaken and grown at 37°C until the optical density at 600 nm reached 0.6. Protein expression was then induced with 1 mM IPTG for 4 hours. The expression level was analyzed using SDS/PAGE (4% stacking gel; 12% running gel). The stacking gel ensures that all proteins arrive at the running gel at the same time, and thus, proteins with the same molecular weight will migrate as tight bands [11]. The non-induced controls (as negative controls) were analyzed in parallel with one another.
2.4. Solubility Testing
The solubility of His-GST-gp46-I(162-214) was checked for expression under appropriate conditions. The E. coli pellets, obtained from 50 mL of culture after IPTG induction, were re-suspended in phosphate-buffered saline (PBS), and samples were subjected to sonication (Bandelin, Germany) on ice. Sonication was performed at a 20 kHz of frequency for 2 minutes, and bacteria were pulsed for 6 × 10 seconds intervals. The cell lysate was then centrifuged at 12,000 rounds per minute (g) and 4°C for 20 minutes. The clear supernatant and pellets were analyzed in 12% polyacrylamide gels using SDS-PAGE.
2.5. Isolation of Inclusion Bodies
After being induced by 1 mM IPTG for 4 hours, the cultures were centrifuged at 5000 g for 10 minutes and harvested. One gram of wet weight cells was washed with 0.9% sodium chloride solution and suspended in 3 mL of lysis buffer (containing 50 mM Tris-HCl, 1 mM EDTA, and 100 mM NaCl; pH = 8.0). Lysozyme was added to a concentration of 100 µg/mL and the cells were incubated on ice for 20 minutes. The lysates were treated with ultrasound (power: 200W; on-time and off-time of the ultrasonic pulse: 2 s) at 4°C for 2 minutes. The soluble cell fractions were removed by centrifugation at 12,000 g and 4°C for 10 minutes. The inclusion bodies were washed twice with 2 mL of washing buffer (containing 50 mM Tris-HCl, 100 mM NaCl, and 0.5% Triton X-100; pH = 8.0), collected by centrifugation at 12000 g and 4°C for 10 minutes, and solubilized in 2 mL of buffer (containing 8 M urea, 50 mM Tris-HCl, and 100 mM NaCl; pH = 8.0) at 4°C for 4 hours.
2.6. Protein Purification
After being centrifugated at 12,000 g and 4°C for 10 minutes, the proteins were applied to the affinity chromatography column. Following extensive washing with binding buffer (containing 20 mM Tris, 500 mM NaCl, and 20 mM imidazole; pH = 8.0), 5 column volumes of elution buffer (containing 20 mM Tris, 500 mM NaCl, and 250 mM imidazole; pH = 8.0) were added to the fusion protein at a flow rate of 1 mL/min. The peak fractions containing the fusion protein were pooled and dialyzed in PBS overnight at 4°C. Bradford protein assay, with bovine serum albumin(BSA) as the standard, were applied to measure protein concentration.
2.7. ELISA
Briefly, 96-well plates were coated with 100 µL of purified protein or synthetic peptide (0 - 25 µg/mL in PBS) and maintained at 4°C overnight. Subsequently, 100 µL of 5% (w/v) nonfat dry milk in PBS was used to block nonspecific binding sites. The blocking continued for 1 hour, and then, the plates were washed with PBST (Tween 20 in PBS). In the next step, 50 µL of infected serum was added to antigen-coated plates in duplicate wells. After incubation at room temperature for 2 hours, the plates were washed and 100 µL of horseradish peroxidase-conjugated gp46-I(162-214) ,HTLV-I peptide were added to each well. Incubation was again performed for 1 hour. After washing, the bound antibody was detected using 100 µL of 3,3′,5,5′-tetramethylbenzidine (TMB) solution. Color development was allowed to proceed for 15 minutes and the reaction was stopped with 50 µL of stop solution. Absorbance was determined at 450 nm using an ELISA reader.
3. Results
3.1. Construction of Expression Vector
For subcloning, restriction enzymes BamHI and EcoRI were added to the 5’ end and 3’ end of the gp46-I(162-214)gene, and the gene was optimized according to E. coli codon usage. After optimization, the codon adaptation index (CAI) was improved to 0.88. CAI values above 0.8 indicate that the gene could be overexpressed in E. coli. Moreover, the GC content of the synthetic gene was increased to 55% which was higher than the average GC content of E. coli (i.e., 50%). The optimized sequence was synthesized and subsequently inserted into the pGS21a vector, moreover, and pGS21a-gp46-I(162-214), containing His-GST-tag for affinity purification, was obtained. The accuracy of subcloning was verified by DNA sequencing. Following these multiple sequence alignments, we detected a sequence similarity between these peptide amino acid residues and the registered sequences (Figure 1).
3.2. Expression of the gp46-I(162-214)
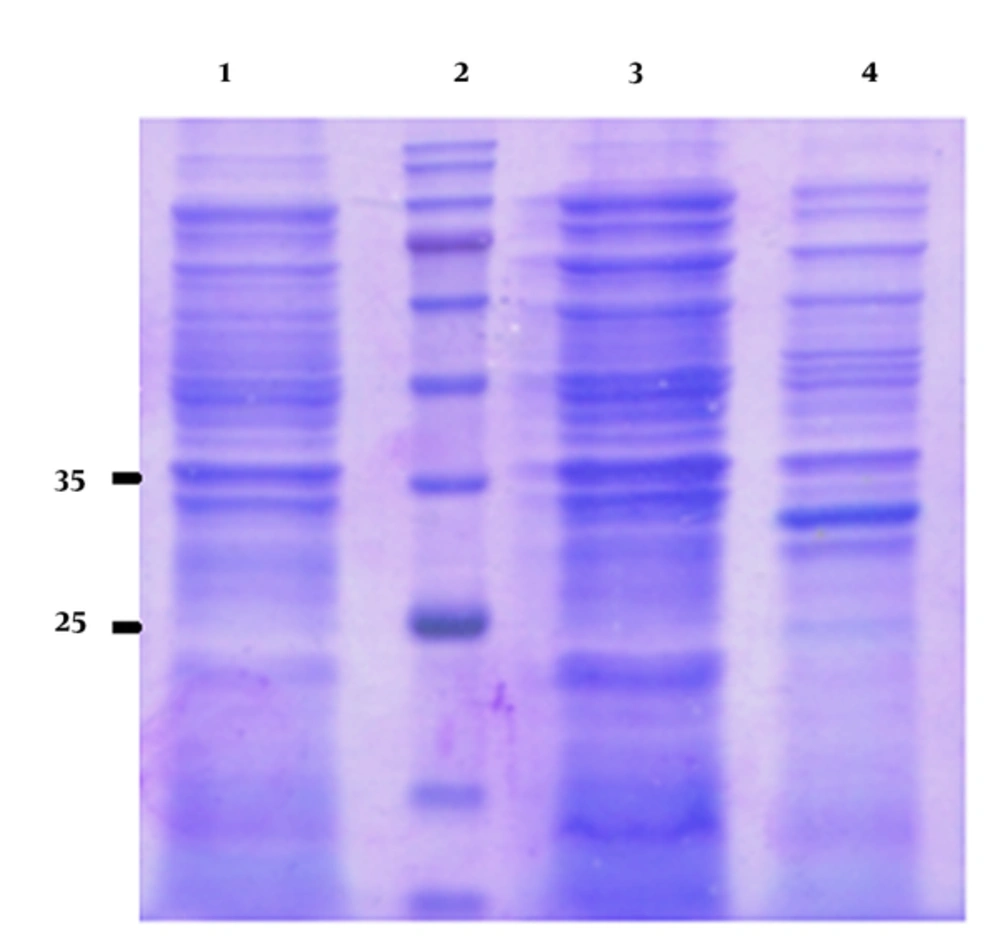
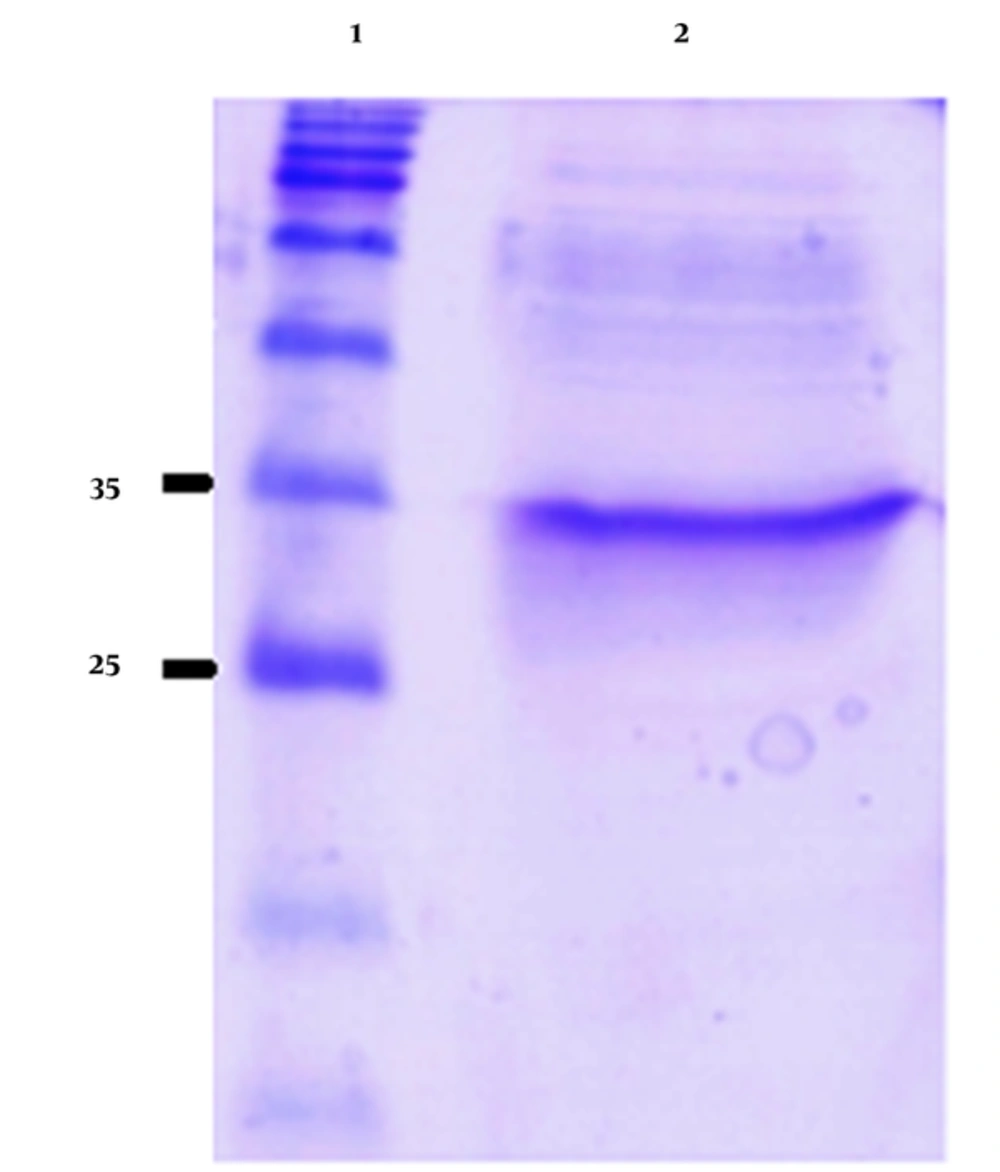
The induced E. coli BL21 (DE3) cell lysate harboring recombinant plasmid was analyzed by SDS-PAGE. The induction time was 4 hours and the IPTG concentration was 1 mM. SDS-PAGE of the lysate of the induced pGS21a-gp46-I(162-214)/BL21 recombinant bacteria revealed a 32-kDa band, corresponding in size to the expected molecular weight of His-GST-gp46-I(162-214) (Figure 1). However, no such band was observed for the non-transformed and non-induced pGS21a/E. coli transformants (Figure 2).
3.3. Solubility Determination
SDS-PAGE of the supernatant and pellet was performed to determine the distribution of the His-GST-gp46-I(162-214) protein in the soluble and insoluble fractions. According to the obtained results, the protein was in the pellet. The results of densitometry and Bradford protein assay revealed that His-GST-gp46-I(162-214) accumulated up to about 30% of the insoluble proteins (Figure 3).
3.4. Purification of His-GST-gp46-I(162-214)
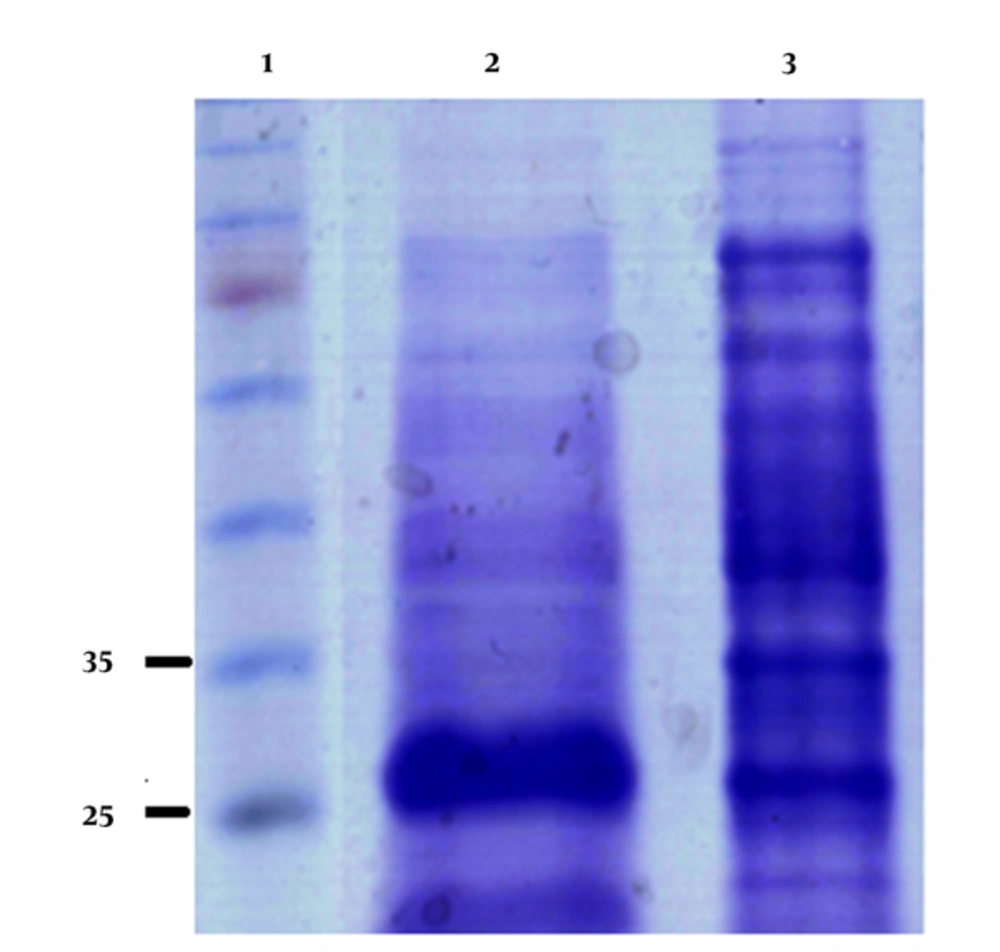
The His-GST-gp46-I(162-214) fusion protein was purified from the solubilized protein aggregates using Ni-NTA affinity chromatography. The proteins without His tags were removed from the Ni-NTA resin using a washing buffer containing 20 mM imidazole. His-tagged His-GST-gp46-I(162-214) was eluted using an elution buffer containing 250 mM imidazole with more than 90% purity (Figure 4).
3.5. ELISA
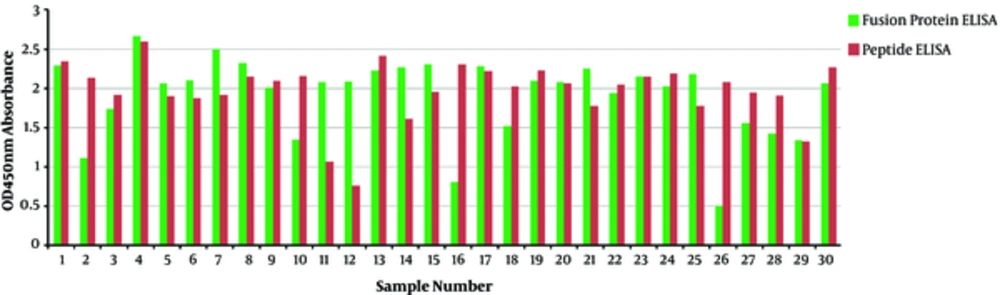
ELISA was used to determine the binding activity of the His-GST-gp46-I(162-214) protein to antibodies present in HTLV-I-infected serum. The results revealed that the His-GST-gp46-I(162-214) protein could specifically bind to anti-HTLV-I antibodies. Moreover, comparison of ELISA results for gp46-I(162-214) peptides and refolded His-GST-gp46-I(162-214) indicated that these interactions were highly specific. According to ELISA results, the peptide showed higher optical density absorbance at 450 nm compared to His-GST-gp46-I (162-214). The high immunodominant epitope present on the plate might have been responsible for this finding (Figure 1).
4. Discussion
In this study, we modified and synthesized the gene encoding HTLV-I gp46-I(162-214) to detect anti-HTLV-1 antibodies in the serum of suspected patients for screening purposes.
HTLV represents a family of human retroviruses with 4 known members and many subtypes [12]. HTLV-I and HTLV-II are major causes of neurologic disorders and lymphocytic leukemia. Peptides derived from these viruses are the most common and most widely used antigens in the serological tests of HTLV infection worldwide [13]. Synthetic peptides are used to detect HTLV-I/-II antibodies and increase its specificity [14].
Protein expression usually leads to the formation of inclusion bodies at 37°C. The key advantage of expression of proteins as an inclusion body is the high level of expression, which reaches to more than 30% of cellular proteins in some cases [15]. The inclusion bodies can be easily isolated considering their differences in size and density before purification; and this can decrease the number of purification steps. However, the refolding of inclusion bodies to bioactive protein is a highly complicated process [16]. Hence, there has been growing interest in soluble expression of inclusion bodies, which is different from peptide expression. In contrast to full length proteins, peptides do not require time-consuming complete refolding to the 3D structure.
In the present study the DNA encoding of gp46-I(162-214) was codon-optimized according to E. coli codon usage because the viral protein sequences contain rare codons for prokaryotic cells. Previous studies reported increased recombinant protein yield following codon optimization [17]. The codon-optimized DNA was subcloned into pGS21a which had an additional His-GST tag to the N-terminus of the protein [4]. The addition of His tag allowed protein purification through a one-step strategy. GST tag is a 26 kDa protein and increases peptide size [18]. This can increase the expression level of the peptide because small peptides expressed in E. coli cells are usually degraded by different proteases present in the host cell. Fusion of peptides with large-sized tags has been found to increase their expression level. GST tag is highly efficient, when positioned in the N-terminal than in the C-terminal of proteins or peptides [19]. Expression of gp46-I(162-214)was induced with IPTG (final concentration 1 mM) at 37°C for 4 hours. Under such conditions, protein expression led to the formation of inclusion bodies. The obtained cells were harvested and lysed by lysis buffer, and the supernatant was discarded. To remove the residual host proteins and DNA, the inclusion bodies were extensively washed in a buffer containing triton, NaCl, Tween-20, and urea. Ultrasound was also used to remove nucleic acids during the isolation of inclusion bodies. Although the washing step is indispensable, the preliminary work can decrease the difficulties and expenses in the next purification procedures and provide a product of high purity. The isolated inclusion bodies were further purified through immobilized metal affinity chromatography (IMAC) which is a highly selective chromatography method widely used for purification. Here, purity more than 90% was achieved as determined by SDS-PAGE after purification. Such a high purity facilitated the next refolding process. Refolding of insoluble recombinant proteins from inclusion bodies generally involves time-consuming processes such as dialysis or dilution. Because considerable amounts of protein are aggregated and precipitated during these processes, refolding was performed at low concentrations to prevent aggregation [20]. After refolding, the protein solution was first dialyzed to eliminate its urea and detergent content and then concentrated. Histidine tagging of recombinant proteins allows on-column refolding and purification to be performed during one single step [21]. Binding of unfolded proteins occurs because the presence of denaturant does not reduce the affinity of the histidine tag for metal ions. In subsequent washing and elution steps, to allow the one-column refolding of the protein, those buffers should be used that do not contain urea. Immobilization of refolded proteins, on a support can minimize protein aggregation and is thus beneficial. Proper refolding of the protein can be confirmed using a functional assay. Results of 30 HTLV-I-infected serum samples by refolded gp46-I(162-214) and synthetic gp46-I(162-214) showed 100% concordance. The results are in agreement with those reported by Yi-ming et al. who found, that the c-terminal half of gp46 can detect 97.6% of the positive samples [22]. In another study, E. coli expression system producing p19 protein has been developed by Mosadeghi et al. In their study, codon optimization and GST fusion resulted in high expression and facile purification of the protein. They reported that this p19 fusion protein reacted to HTLV-I antibodies and showed strong sensitivity and specificity and was compatible to the ELISA results of p19 [4], which is similar to our results. In the present study, no cross reactivity was detectable in sera from healthy controls. These data indicated that this experimentally designed antigen showed good specificity.
4.1. Conclusions
In this study, we developed a facile method and presented a strategy for high level expression, purification, and refolding of gp46-I(162-214) to be used in ELISA for screening the suspected cases of HTLV-1 infection. Recombinant His-GST-gp46-I(162-214) was purified and refolded using IMAC. Moreover, compared to long and time- consuming purification procedures that require dilution, dialysis, and concentration, the proposed approach reduced the purification and refolding time by integrating all processes in 2 steps. One limitation of this study was that the number of studied HTLV patients was low; and thus, the serodiagnosis might have been under estimated so it should be further investigated in future studies.