1. Background
Aflatoxins, a polyketide group derived from furanocoumarins, are highly toxic carcinogens produced by Aspergillus flavus and other closely related species (1). The four major types of aflatoxins are B1, B2, G1, and G2. Aflatoxin B1 is more important because it is more toxic and more carcinogenic (2). Aflatoxin contaminated foods can lead to acute hepatitis, immune suppression, liver cancer, and neutropenia (3). A collection of 25 genes within a 75-kb region of genome A. flavus are involved in aflatoxin production (1, 4). Studies have shown that there is a high correlation between the production of aflatoxin and the expression of aflQ, aflO, aflD, aflS, and aflR genes in A. flavus (5, 6).
Biological and chemical control of pistachio A. flavus is restricted or prohibited due to international rules and regulations (7). Therefore, studies have focused on the use of natural compounds, especially essential oils and plant extracts, to prevent the growth of these fungi and the production of aflatoxin (8-15). Pasture plants, Capparis spinosa, Withania somnifera, and Peganum harmala have been proved to have antifungal effects. Studies have shown that W. somnifera extracts have antifungal properties against Fusarium oxysporum and Colletotrichum capsici (16, 17), and methanolic extract of W. somnifera has fungicidal properties against A. flavus and A. niger (18, 19). Furthermore, the ethanolic extract of W. somnifera has fungicidal properties against Candida albicans (20). Capparis spinosa has antifungal properties (21-23). Studies have shown that P. harmala extract has fungicidal properties (24-27).
2. Objectives
The purpose of this study was to investigate the effect of extracts of some pasture plants on A. flavus growth and expression of some major genes in aflatoxin biosynthetic pathway.
3. Methods
The present experimental study was conducted at the agricultural research institute, University of Zabol and approved by the ethical committee of the Islamic Azad University of Zahedan.
3.1. Plant Material and Preparation of Plant Extracts
Plants C. spinosa, W. somnifera, and P. harmala were collected from Sistan and Balochestan province, Iran. The collected plants were identified at the department of Medicinal plants of agricultural research institute, University of Zabol. The seeds of P. harmala and leaves of C. spinosa and W. somnifera were ground using a mortar and pestle. Ten grams of grinded powder was soaked in 100 mL of 96% ethanol for one day. The supernatant was filtered through Whatman No. 1 filter paper. The dried extracts were preserved at 4°C for further use (28).
3.2. Minimal Inhibitory Concentration (MIC) and Minimum Fungicidal Concentration (MFC)
The MIC was determined by a microplate method (29). The MFC values of plant extracts were determined using the method described by Espinel-Ingroff et al. (30).
3.3. Effect of C. spinosa Extract on Aflatoxin Biosynthesis-Related Genes
The standard isolate of A. flavus PFCC 116 was cultured in potato dextrose broth (PDB, Sigma-Aldrich, Germany) (as control) and PDB medium containing 200 mg mL-1 of C. spinosa extract and incubated at 35°C for three days. After three days of incubation, the fungal mycelia was harvested. Total RNA was extracted using the Top plant and fungi RNA purification kit (TOPAZ GENE RESEARCH, Iran), based on the method described in the kit's manufacturer's instructions. The first strand of cDNA was synthesized using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, Waltham, Massachusetts, U.S.A.). Expression of genes was evaluated using the quantitative real-time PCR (qRT-PCR) technique with Eva Green fluorescent dye and their specific primers. The qRT-PCR was performed with a Rotor-Gene 3000 (Corbett Robotics, Australia). The process was performed with 20 µL of reaction mixture consisting of 1 × HOT FIREPol® EvaGreen® qPCR Mix Plus (Solis BioDyne, Riia, Estonia), 1 µL cDNA, and 10 µM of each forward and reverse primer. The following protocol was applied: initial polymerase activation: 15 minutes at 95°C; 40 cycles of 30 seconds at 95°C, 30 seconds at 61°C, and 45 seconds at 72°C. The characteristics of the primers used for qRT-PCR are listed in Table 1. The relative changes in the expression of target genes were normalized to the β-tubulin housekeeping gene (31). Analysis of relative gene expression data was carried out using the Relative Expression Software Tool (REST) 2009 software version 2.0.13 (Qiagen, Hilden, Germany) (32).
| Gene | Forward Primer (5' - 3') | Reverse Primer (5' - 3') | Amplicon Length, bp | Tm, ºC | Accession Number |
|---|---|---|---|---|---|
| aflR | CTCAAGGTGCTGGCATGGTA | CAGCTGCCACTGTTGGTTTC | 86 | 61 | XM-002379905 |
| aflQ | GTGTCCGCAGTGTCTAGCTT | GCTCAAAGGTCGCCAGAGTA | 115 | 61 | XM-002379890 |
| aflO | GGGACGATTTGATGGAGCAG | TTAGGTTCTCTTGGCTACAGCA | 101 | 61 | XM-002379892 |
| aflS | AAGCTAAGGCCGAGTCTGG | TGCTCGCTAGTTTCAAGGCA | 75 | 61 | XM-002379904 |
| β-tubulin | CTTGTTGACCAGGTTGTGGAT | GTCGCAGCCCTCAGCCT | 51 | 61 |
Nucleotide Sequences of Primers Used in This Study
3.4. Statistical Analysis
In this experiment, three replicates were considered for each treatment and the meanings were compared with t-test using SPSS statistics version 19 (IBM, U.S.A). The differences with P < 0.05 were considered significant.
4. Results
4.1. Antifungal Activity
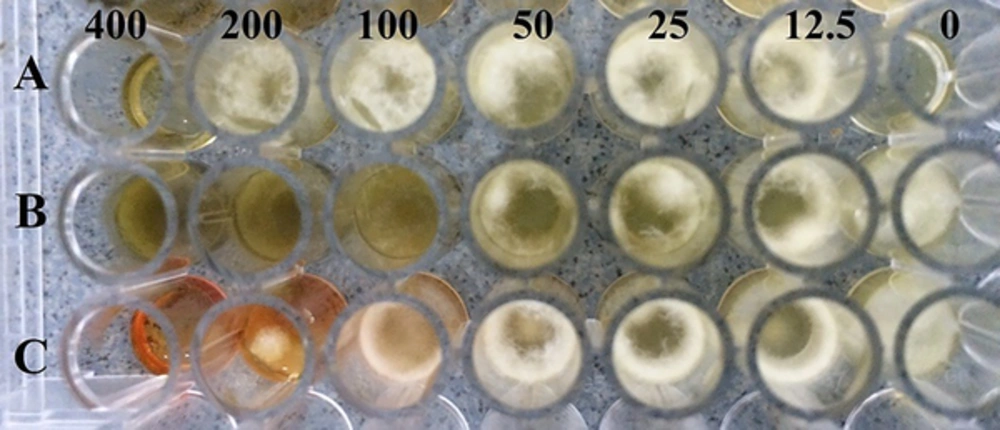
The results of the effects of different concentrations of C. spinosa, W. somnifera, and P. harmala extracts on A. flavus PFCC 116 growth two days after incubation at 35°C is shown in Figure 1. Aspergillus flavus covered the entire surface of the control microplate and in other microplates, the fungal growth rate decreased with increasing concentration of extracts, and in comparison with other microplates, the control microplate had the ability to inhibit growth of A. flavus by the extracts. The MICs of C. spinosa, W. somnifera and P. harmala extracts were 200, 400, and 400 mg/mL, respectively (Table 2). The MIC values ranged from 200 to 400 mg/mL, and the C. spinosa extract had the lowest MIC value (200 mg/mL), and W. somnifera and P. harmala extracts had the highest MIC value (400 mg/mL).
| Plant Species | MIC | MFC |
|---|---|---|
| Withania samnifera | 400 | - |
| Capparis spinosa | 200 | 400 |
| Peganum harmala | 400 | - |
Minimal Inhibitory Concentration (MIC) and Minimum Fungicidal Concentration (MFC) of Crude Extracts from Plant Species Against Aspergillus flavus
Fungicidal effects of different concentrations of C. spinosa, W. somnifera, and P. harmala extracts on A. flavus is shown in Table 2. The MFC of C. spinosa extract was 400 mg/mL and the extracts of W. somnifera and P. harmala had no fungicidal activity, and the C.spinosa extract had the lowest MFC value (400 mg/mL) and had the highest fungicidal effect on A. flavus.
4.2. Effect of C. spinosa Extract on Expression of Major Genes in aflatoxin Biosynthetic Pathway
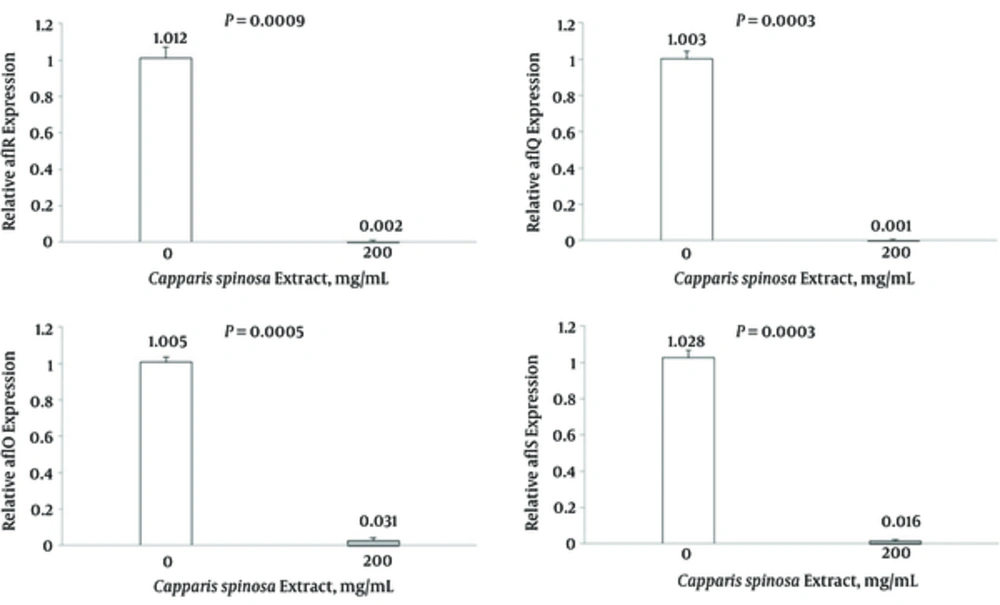
Results of expression of aflR gene in samples treated with C. spinosa extract (200 mg/mL) and non-treated (control) A. flavus PFCC 116 are shown in Figure 2. After treatment with C. spinosa extract, a strong reduction (98.8%) was observed at the transcriptional level of aflR gene. The results showed significant differences between the expression of aflR gene expression in treated and non-treated samples at 5% level. Results of determination of expression of aflQ gene in samples treated with C. spinosa extract (200 mg/mL) and non-treated (control) A. flavus PFCC 116 are shown in Figure 2. After treatment with the C. spinosa extract, a strong reduction (99.9%) was observed at the transcriptional level of the aflQ gene. The results showed significant differences between the expression of aflQ gene expression in treated and non-treated samples at the 5% level. Results of determination of expression of aflO gene in samples treated with C. spinosa extract (200 mg/mL) and non-treated (control) A. flavus PFCC 116 are shown in Figure 2. After treatment with the C. spinosa extract, a strong reduction (96.9%) was observed at the transcriptional level of the aflO gene. The results showed significant differences between the expression of aflO gene expression in treated and non-treated samples at the 5% level. Results of determination of expression of aflS gene in samples treated with C. spinosa extract (200 mg/mL) and non-treated (control) A. flavus PFCC 116 are shown in Figure 2. After treatment with the C. spinosa extract, a strong reduction (98.4%) was observed at the transcriptional level of the aflS gene. The results showed significant differences between the expression of aflS gene expression in treated and non-treated samples at the 5% level.
Level of mRNA expression of aflR, aflQ, aflO and aflS in the treated (200 mg mL-1) and the non-treated (control) samples in Aspergillus flavus. Each sample is normalized for the amount of the template to the levels of β-tubulin. Significant differences were observed for mRNA levels for each of gene between the treated and the non-treated fungal cell (P < 0.05). The bars represent the mean ± SD values.
5. Discussion
In this research, the antifungal properties of the ethanol extract of C. spinosa were investigated against a toxinogen isolate of A. flavus and its effect on the reduction of the expression of the main genes in aflatoxin biosynthetic pathway, aflR, aflQ, aflO and aflS genes. According to several reports, C. spinosa had antifungal activity at certain concentrations. Mahboubi and Mahboubi (21) investigated antifungal activity of ethanolic extract of root and fruit of C. spinosa against A. niger, A. parasiticus, and A. flavus. The ethanolic extract of root and fruit of C. spinosa showed fungicidal and inhibitory effects against A. niger, A. parasiticus and A.flavus, which was similar to the results of the current research. Ali-Shtayeh and Abu Ghdeib (22) examined the antifungal properties of aqueous extract of C. spinosa against Microsporum canis, Trichophyton mentagrophytes and Trichophyton violaceum. The aqueous extract of C. spinosa completely prevented the growth of M. canis and T. violaceum. Al-Askar (23) examined the antifungal properties of ethanolic extract of C. spinosa in vitro against Alternaria alternata, F. oxysporum, Phoma destructiva, Rhizoctonia solani and Sclerotium rolfsii. The C. spinosa extract had different degrees of antifungal properties against the tested fungi.
In this study, the C. spinosa extract reduced expression of aflR, aflQ, aflO and aflS genes. The aflR gene is required for transcriptional activation of most structural genes in A. flavus (1). The aflS gene enhances the transcription of toxigenic genes in A. flavus and the aflO and aflQ genes, producing O-Methyltransferase B and Cytochrome P450 monooxygenase enzymes, are needed for biosynthesis of aflatoxins (1). Verheecke et al. (33) examined the expression of aflatoxin genes during the interaction of Streptomyces-Aspergillus. Expression of aflR and aflS was generally repressed in A. flavus and A. parasiticus. Al-Saad et al. (34) investigated the effect of four bacteria antagonists against A. flavus to control the production of aflatoxin B1. These studies showed that some of the bacterial species could significantly inhibit the relative expression of the aflD and aflR genes. Mohseni et al. (12) studied the effect of Glycyrrhiza glabra extract on aflR gene expression and aflatoxin production in A. parasiticus. Five hundred mg/mL concentration of G. glabra extract inhibited aflR gene expression and consequently the aflatoxin production efficiently in A. parasiticus. Jahanshiri et al. (35) studied Eugenol effect on A. parasiticus growth and aflatoxin production and the expression of major genes involved in the aflatoxin biosynthesis pathway. Eugenol (62.5 and 125 µg mL-1) significantly suppressed the expressions of ver-1, nor-1, pksA, omtA, and aflR genes. Jahanshiri et al. (36) examined the effect of curcumin on fungal growth and aflatoxin production in A. parasiticus NRRL 2999. The curcumin significantly inhibited the expression of the aflR gene.
5.1. Conclusion
In general, the C. spinosa extract can have fungicidal and inhibitory effects on A. flavus and reduce expression of major genes in aflatoxin biosynthetic pathway and further studies are proposed because this study is the first major study on the effect of the C. spinosa extract on expression of major genes in aflatoxin biosynthetic pathway.