1. Background
Vibrio vulnificus strains are halophilic, motile, curved, gram-negative bacteria that exist in coastal waters worldwide and are a commensal bacteria in shellfish and fish (1). The bacteria is positive oxidase (2). The strains of V. vulnificus are divided to three biotypes. All biotypes of V. vulnificus are pathogenic for humans. Biotype 1 is the most prevalent cause of septicemia (3). The number of culturable V. vulnificus strains typically decreases by reducing water temperature. In this condition (water temperature below 15°C), the bacteria may enter a viable but non-culturable (VBNC) step. The highest densities are observed during the warm months. Salinity influences the numbers of culturable V. vulnificus, as the highest densities are generally detected at salinities between 5% and 25% (4). Roland et al. introduced the first clinical case of V. vulnificus infection in 1970 (5). In addition, the first wound infection created by V. vulnificus was reported in 1979 by Blake et al. (6). Ingestion of raw or inadequately cooked seafood containing V. vulnificus can lead to sepsis. The signs and symptoms of sepsis caused by these bacteria involve fever, chills, nausea, endotoxic shock, and the development of secondary wounds on the extremities of patients. Mortality rates have been reported as greater than 50% in patients with primary sepsis, and about 25% for wound infections. Wound infections progress quickly to cellulitis, ecchymoses, and bullae, which can lead to necrotizing cellulitis and even secondary sepsis at the site of infection (7, 8). The virulence factors of V. vulnificus include resistance to stomach acid, capsule, endotoxin, cytotoxins, pilus, and flagellum (8, 9). Several PCR assays have been designed for diagnosis of V. vulnificus yet were unable to find V. vulnificus strains. Therefore, the gyrB gene was chosen for detection of V. vulnificus as previously described by Kumar et al. (10). This gene codes the B subunit of the gyrase, which removes positive supercoils during DNA replication (10). Up to now, no investigation has been done on the prevalence of the bacteria in coastal water of Babolsar. Babolsar is one of the coastal cities of the Mazandaran province (north of Iran). The goal of this investigation was to define the prevalence of V. vulnificus in seawater samples, collected from coastal waters of Babolsar by the PCR method.
2. Methods
2.1. Isolation of Vibrio vulnificus
Three hundred (300) coastal seawater specimens were taken during the spring and summer of 2017. The sample size was determined by the Morgan table. The reason to choose spring and summer for the study was that increasing temperature leads to higher salinity water; therefore, bacteria isolation will be possible. Seawater samples were added, at a depth of 50 cm, to sterile bottles. In sampling, water temperature was measured using a thermometer. Samples were processed within six hours of sampling. To isolate V. vulnificus from seawater, 25-mL samples were added to 225 mL of peptone water and kept at 37°C for six hours. Ten microliters of the cell suspension were then transferred onto thiosulfate citrate bile sucrose (TCBS) agar and incubated at 37°C for 24 hours. Suspected colonies of V. vulnificus (green colonies) were picked from the plates and identified by PCR (11, 12).
2.2. DNA Extraction
DNA extraction was accomplished by the boiling method. In total, 1.5 mL of an overnight culture of V. vulnificus was centrifuged at 6000 rpm for five minutes. The cell pellet was suspended in 300 μL of TE buffer (10 mM Tris-HCl, 0.5 mM EDTA, pH 8), boiled for 10 minutes, and subsequently cooled on ice for five minutes. After incubation on ice, the tube was centrifuged at 10,000 rpm for 5 minutes at 4°C. The supernatant (200 μL) was added to a fresh sterile tube and stored at -20°C for long preservation (13).
2.3. Confirmation of Vibrio vulnificus by the Polymerase Chain Reaction
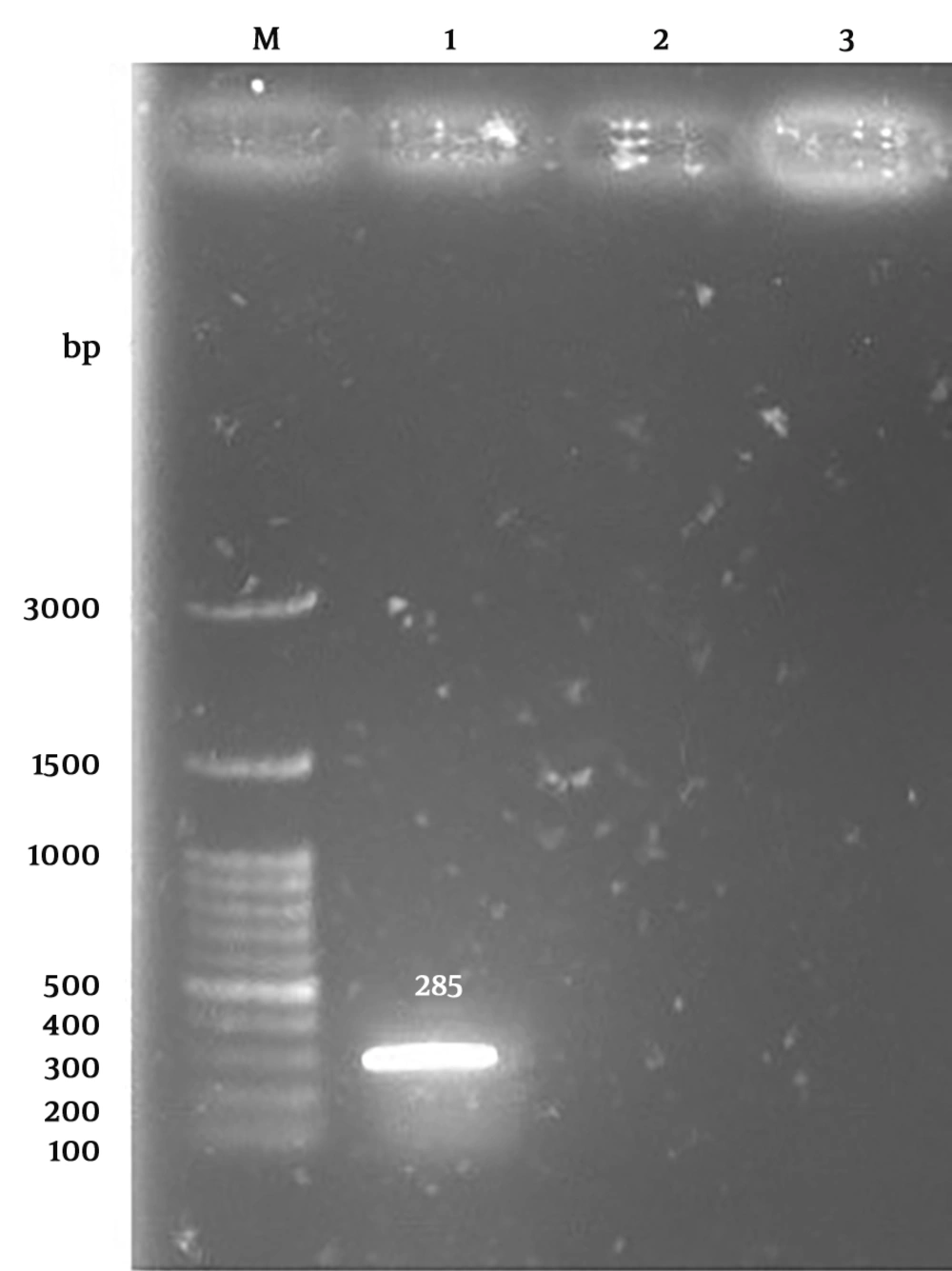
Confirmation of all V. vulnificus strains was performed on all of the green colonies that appeared on TCBS Agar by the PCR technique, as formerly reported by Kumar et al. in 2006. Due to unavailability of a positive control, detection was done via sequencing of PCR products. The PCR products were sequenced by Bioneer corporation. The primers were used for the amplification of a 285-base pair (bp) part of V. vulnificus gyrB gene (10). Table 1 shows the sequence of primers employed for the current investigation.
| Target Organism | Target Gene | Primer Sequence (5′ - 3′) | Size of Product (bp) | Reference |
|---|---|---|---|---|
| V. vulnificus | gyrB | F-GTCCGCAGTGGAATCCTTCA | 285 | Kumar et al, 2006 (10) |
| R-TGGTTCTTACGGTTACGGCC |
The Sequence of Primers Used in the Current Investigation
The PCRs were performed in a final volume of 20 μL. The reaction mixture included 2 μL of DNA template, 2 μL of 10x PCR buffer, 0.4 μL of deoxynucleoside triphosphates (10 mM), 0.6 μL of 50 mM MgCl2, 0.2 μL of Taq DNA polymerase (5 U), 0.5 μL of each primer (20 pmol), and 10.8 μL of distilled water. Initial denaturation at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds. The final extension was done at 72°C for five minutes (12). Gel electrophoresis of PCR products was accomplished on a 2% agarose gel and observed by UV transilluminator after ethidium bromide (EtBr) staining. Data were analyzed by the SPSS software version 16.
3. Results
Vibrio vulnificus creates green colonies on the TCBS agar. Of 300 seawater samples, 276 V. vulnificus presumptive green colonies were grown on TCBS agar plates. Gel electrophoresis of PCR products generated a 285-bp segment corresponding to the gyrB gene (Figure 1).
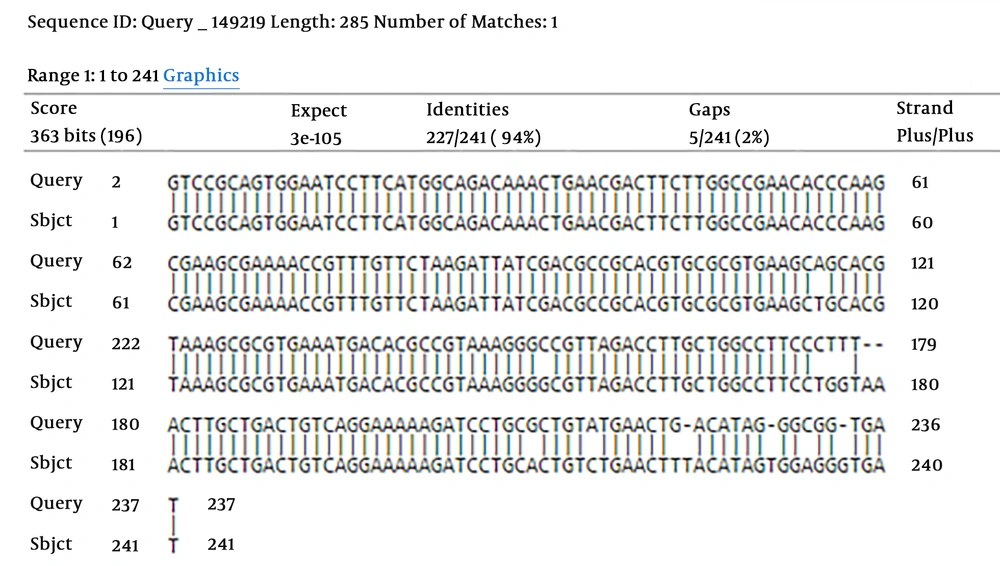
To establish the accuracy of the primers, the PCR product was sequenced using forward and reverse primers. Thereafter, the sequences were aligned with the corresponding region of nucleotide sequence, accessible on the GenBank database with accession number: NC_005139.1. The basic local alignment search tool (BLAST) analysis revealed that the PCR product derived from the V. vulnificus sequence showed similarity of 94% with the GenBank gyrB sequence. Therefore, V. vulnificus was detected (Figure 2).
The results of the PCR technique showed two (0.72%) positive amplifications of the gyrB gene from 276 the presumptive V. vulnificus isolates. Vibrio vulnificus was not detected in any specimens collected during March, April, and May (average water temperature of 15°C) yet was observed in water samples collected during summer (average water temperature of 25°C).
4. Discussion
Vibrio vulnificus enters the body via the consumption of seafood or through an open wound exposed to seawater, contaminated with the bacteria. Due to its high proliferation, wound infections may quickly progress to necrotizing fasciitis and septicemia (14). The investigation was done to gain public awareness of bacteria existence in the area under study. This study was the first investigation in relation to the isolation and molecular detection of bacteria in the coastal seawater of Babolsar, northern Iran. Shaw et al. (15) reported that V. vulnificus strains were detected from the total sampling sites in the Chesapeake Bay and Maryland Coastal Bays. In a study performed by Paydar and Thong (12), a prevalence of 23% of V. vulnificus was reported from seawater in Kuala Lumpur, Malaysia. In another study conducted by Rodgers et al. (16) in Colombia, V. vulnificus was detected in 11% of seawater samples. Maugeri et al. (17) showed low incidence of V. vulnificus strains in the Mediterranean Sea. There are reports of the isolation of V. vulnificus strains from fish, shrimp, and crab in the Persian Gulf (18). In current study, two (0.72%) isolates were identified as V. vulnificus strains by the PCR method. Different results could be due to the levels of contamination of area under study. Furthermore, because of low salinity of Caspian Sea, isolation of the bacteria was lower than other studies performed in other seas. The abundance of V. vulnificus in coastal waters was highly correlated with the water temperature. The isolation of culturable forms was done in the warm months of the summer. Maugeri et al. isolated V. vulnificus from the water at the temperature range of 20 to 21.5°C (17). In this study, the bacteria were isolated in summer (mean water temperature of 25°C). It seems that the findings could show the association of the culturable forms with high water temperature. In the current research, V. vulnificus was not isolated from specimens obtained during spring. In the Gulf of Mexico, V. vulnificus was not isolated when seawater temperature was under 18°C (19). In the study conducted by Pfeffer et al., the bacteria were not isolated at water temperatures below 14°C (20). When the water temperatures reach below 13°C, V. vulnificus strains enter the VBNC state, in which bacteria are metabolically active yet don’t grow on conventional culture media (21, 22). In conclusion, due to the presence of V. vulnificus in the area under study, the bacteria may be dangerous to swimmers, who have open wounds. Of the limitations of the present research were the high mortality rate of the bacteria. Moreover, the sampling should be performed in the warm seasons, when the water temperature exceeds above 20°C. Regarding the transmission of V. vulnificus, which can occur via fish consumption, it is recommended that a study should be carried out on the presence of the bacteria in fish of the Caspian Sea.