1. Background
The human intestine, as a vital anatomical location, has the largest number of microbes; thus, it is not hard to predict the leading role of the microbes as well as the bacteria in health status and different colon diseases in this organ, especially in the development of colorectal cancers (1). Several clinical studies have demonstrated a strong association between invasive infections with Streptococcus gallolyticus and colon neoplasia in humans (2). Colorectal cancer is the fourth most common cancer in the world (3). The incidence of this cancer varies in different areas. In the United States and England, colorectal cancer is the second most common cancer after breast cancer in women and prostate cancer in men (4, 5). In Iran, the number of patients developing colorectal cancer is increasing, and it is prevalent in both genders (6). The colon is a hollow muscular tube that starts from the end of the small intestine or ileum and ends in the anus. The length of the intestine is 1.5 m, and its largest diameter is reported to be at the beginning of the large intestine. Its diameter decreases to the rectum (7). The human's intestine is naturally exposed to 1014 microorganisms, which can be hazardous to people's health and cause intestinal diseases (8). Therefore, identifying certain microorganisms enables us to improve our knowledge for the diagnosis, prevention, and treatment of these cancers (9-12). S. gallolyticus (Streptococcus bovis biotype I) and Helicobacter pylori can cause colorectal cancer (13). Streptococcus gallolyticus subsp. gallolyticus (Sg), formerly known as the S. gallolyticus, is an opportunistic human pathogen that causes bacteremia and endocarditis. This pathogen stimulates cancer cells through the β-catenin signaling pathway (14, 15), and naturally lives in the human gastrointestinal tract. About 2.5% to 15% of healthy people bear this pathogen (16, 17). During the last four decades, the relationship between bacteria and colorectal cancer has been studied using serologic testing. However, the molecular findings have raised controversies. All of these studies have demonstrated that intestinal bacterial infection is associated with an increased risk of colorectal cancer and is mainly related to a bacterial agent, including viruses, bacteria, and parasitic agents that may contribute to colorectal cancer (12). Polymerase chain reaction (PCR) is the basis for detecting S. gallolyticus genome in colorectal cancer, in which the tissue is detached by colonoscopy. A part of the bacterial gene is identified, and the relevant primers are produced, which confirm the final diagnosis for existing bacteria inside the colon. Moreover, culturing this Gram-positive bacterium and comparing it with the molecular method provide us with a better diagnosis (18). Recent advances in bacterial systematics have allowed for the use of the 16S ribosomal RNA (rD A) gene as a target for the detection and identification of various microorganisms. The present report describes the development of specific primers that target the 16S rD A gene to rapidly identify and differentiate human and ruminal strains of S. gallolyticus (19).
However, a recent study conducted in Germany using sensitive PCR to detect S. gallolyticus indicated a higher carriage rate estimated at 62.5% in the stools of 99 healthy volunteers (20). So far, no study has been performed to detect the colonization of S. gallolyticus of the large intestine directly in colorectal cancer tumors using advanced PCR molecular testing in northern Iran.
2. Objectives
The current study aimed at identifying the molecular structure of S. gallolyticus bacteria in patients with colon diseases.
3. Methods
This descriptive research was conducted on 55 tissue samples of people with colon diseases undergoing colonoscopy in two hospitals of Babol and Chalous for eight months from February 2017 to September 2018. Written informed consent was obtained from patients prior to sampling. A checklist was used to collect data, including demographic information, such as age, sex, duration of illness, place of residence, family history of the disease, and having clinical symptoms, such as abdominal pain, rectal bleeding, fatigue, history of rectal hemorrhage, history of hemorrhoids, constipation, diabetes, and colonoscopy findings (polyp size, site, and histopathology). Patients with missing clinical information and incomplete colonoscopy examination were excluded. The tissue samples were collected by colonoscopy by a specialist physician for DNA extraction and performing PCR using a microtube (2 cc) containing sterile physiology serum to be sent to the molecular laboratory at -20°C.
3.1. Polymerase Chain Reaction
For DNA extraction, 25 mg of tissue from each sample was dried with a few drops of liquid nitrogen. DNA extraction from the chopped tissue was performed according to the instruction of the FAVORGEN kit (Taiwan) by column method using FATG2 buffers, K proteinase, and ethanol. Ultimately, to dissolve the extracted DNA, the Elution Buffer available in the kit was used. The DNA extraction was performed quantitatively (1.6 < OD < 1.9) and qualitatively using PCO3 (5'-ACACAACTGTGTTCACTAGC-3') and PCO4 (5'-CAACTTCATCCACGTTCACC-3') primers, which duplicate a part of the human β-globulin gene.
The primers were developed by Primer-Blast from NCBI and Primer3 software and synthesized by the Sinoclon company, and received as lyophilized.
Using 5'-CAATGACAATTCACCATGA-3' and 5'-TTGGTGCTTTTCTCTTGTG-3' primers, the amplification and identification of the considered genes were performed. After preparing the final volume of each PCR reaction (20 μL), the vials were placed in the thermocycler machine. The temperature and time condition for a PCR reaction for the genes were performed. Finally, the detection and photographing of PCR products under UV light after electrophoresis of the products for 2 to 4 minutes with agarose gel containing 1 - 0.5 μg/mL of Ethidium bromide were performed (Figure 1).
3.2. Statistical analysis
The results were analyzed by SPSS 16 software and Fisher test for the analysis of PCR results. A P < 0.05 was considered significant.
4. Results
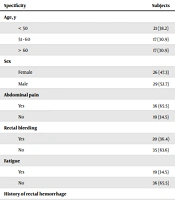
In this descriptive study, 55 patients with a mean age of 52.7 ± 13.9 years were included. The demographic characteristics and distribution of S. gallolyticus in culture samples are presented in Table 1, according to the clinical symptoms and characteristics of the patients. Table 1 signifies a significant relationship between family history of colorectal cancer (P = 0.011) and diabetes, (P = 0.008) and frequency of S. gallolithic.
| Specificity | Subjects | Positive Cases | Negative Cases | P Value |
|---|---|---|---|---|
| Age, y | 0.999 | |||
| < 50 | 21 (38.2) | 3 (14.3) | 18 (85.7) | |
| 51 - 60 | 17 (30.9) | 3 (17.6) | 14 (82.4) | |
| > 60 | 17 (30.9) | 3 (17.6) | 14 (82.4) | |
| Sex | 0.572 | |||
| Female | 26 (47.3) | 4 (15.4) | 22 (84.6) | |
| Male | 29 (52.7) | 5 (17.2) | 24 (82.8) | |
| Abdominal pain | 0.330 | |||
| Yes | 36 (65.5) | 7 (19.4) | 29 (80.6) | |
| No | 19 (34.5) | 2 (10.5) | 17 (89.5) | |
| Rectal bleeding | 0.577 | |||
| Yes | 20 (36.4) | 3 (15.0) | 17 (85.0) | |
| No | 35 (63.6) | 6 (17.1) | 29 (82.9) | |
| Fatigue | 0.373 | |||
| Yes | 19 (34.5) | 4 (21.1) | 15 (78.9) | |
| No | 36 (65.5) | 5 (13.9) | 31 (86.1) | |
| History of rectal hemorrhage | 0.390 | |||
| Yes | 8 (14.5) | 2 (25.0) | 6 (75.0) | |
| No | 47 (85.5) | 7 (14.9) | 40 (85.1) | |
| History of familial colorectal cancer | 0.011 | |||
| Yes | 11 (20.0) | 5 (45.5) | 6 (54.5) | |
| No | 44 (80.0) | 4 (9.1) | 40 (90.9) | |
| History of hemorrhoids | 0.214 | |||
| Yes | 8 (14.5) | 0 (0.0) | 8 (100.0) | |
| No | 47 (80.5) | 9 (19.1) | 38 (80.9) | |
| Constipation | 0.572 | |||
| Yes | 26 (47.3) | 4 (15.4) | 22 (84.6) | |
| No | 29 (52.7) | 5 (17.2) | 24 (82.8) | |
| Diabetes | 0.008 | |||
| Yes | 20 (36.4) | 7 (35.0) | 13 (65.0) | |
| No | 35 (63.6) | 2 (5.7) | 33 (94.3) | |
| Diagnosis of the disease | 0.186 | |||
| 1 month ≥ | 10 (18.2) | 0 (0.0) | 10 (100.0) | |
| 1 month - 1 year | 22 (40.0) | 3 (13.6) | 19 (86.4) | |
| ≤ 1 year | 23 (41.8) | 6 (26.1) | 17 (73.9) |
Demographic Characteristics and Frequency of Streptococcus gallolithic Bacteria in Culture Samples a
From 55 biopsy samples of patients with colon diseases, 3 samples (5.5%) were positive and 52 cases (94.5%) were negative for the presence of S. gallolyticus DNA. Fisher’s exact test showed a significant correlation between the prevalence of S. gallolyticus and colon diseases (P = 0.015). Eleven cases (20%) had a history of familial colorectal cancer, of whom 5 cases (45.5%) were observed with the bacteria (P = 0.011). In the analysis of PCR gels, a band of 406 bp after the PCR process showed that all of the samples were in a favorable condition considering the quality of the extracted DNA. The presence of different bands in the sample indicates the presence of S. gallolyticus bacteria. The electrophoresis of PCR products was done on agarose gel 1.5% (Figure 1).
5. Discussion
Laboratory findings of this study on PCR revealed that of 55 samples of biopsy tissues (polyps, colitis, cancer, and inflammation of the intestines), 5.5% were positive for S. gallolyticus. In a study by Sarokhani et al., PCR showed the presence of bacterial genomes in 36 positive samples out of 100 samples (21). A study by Gilermo on 568 patients over 65 years of age identified 15 cases with S. gallolyticus bacteria (22). In another study, Pui-ying et al. discovered that out of 537 patients, 12 positive samples (2.3%) were positive using PCR, and 15 cases were positive after culturing (23). Therefore, the positive predictive value for the diagnosis of S. gallolyticus bacteria is higher using the molecular method. Rhoads et al. found that the majority of bacteria that were recognized by the molecular method were not detectable by the culture method (24, 25). Other studies also showed that the PCR method had a higher sensitivity for bacteria diagnosis (26, 27).
In our study, 11 cases (20%) had familial colorectal cancer, of whom 5 cases (45.5%) had the bacteria (P = 0.011). Furthermore, Norfleet and Mitchel reported a positive prevalence of S. gallolyticus 3% in the biopsy samples of colorectal cancer, 2.5% in normal tissue of the intestine, and 0.0% in polyp samples (28). Using frequency-based molecular techniques, Abdulamir et al. found 48.7% of DNA sequences of S. gallolyticus in samples of patients with colorectal cancer in contrast to 4% of normal samples (29). Other studies have shown that S. bovis is associated with colorectal adenocarcinoma, especially in female patients, which contradicted our findings (30). According to the mentioned studies, the association between S. gallolyticus and colorectal cancer is still controversial. The differences in results can be traced to the genetic background as well as the geographical differences of the patients.
On the other hand, 20 out of 55 subjects (36.4%) had diabetes, of whom 7 subjects (35%) had S. gallolyticus bacteria (P = 0.008). In a study by Zammit in 2013 on patients suffering from colon diseases, it was realized that 26% of the subjects had diabetes in addition to S. gallolyticus infection (31). In a case-control study, Ellezin reported a Streptococcus infection in a diabetic patient who had colon cancer (32).
5.1. Conclusion
Molecular techniques have created a highly sensitive antibacterial framework bed for the detection of microbial pathogens. The sensitivity and specificity of PCR depend on the primer used. However, one of the most important factors in using PCR is the infections that can occur during DNA extraction, which is the preparation of the reaction mixture and the extension. Nonetheless, considering the advantages and disadvantages as well as the characteristics of this method, at present, this method can be considered comprehensive and standard. Thus, the simultaneous use of this method is recommended whenever a quick result is sought, or there is a suspicion of sample infection and late-growing microorganisms.
5.2. Limitation and Suggestion
Due to the time limit of more than 50 cases, access to the samples of people with colon cancer and other clone diseases was not possible. We suggest further investigations to confirm the role of S. gallolyticus in the development of colon cancer in the future using a larger sample size (patients) compared with healthy controls.