1. Background
Leishmaniasis as a parasitic disease is now considered as a neglected disease by WHO in many countries around the world. However, the high incidence rate of the disease occurs in tropical and temperate regions where the Leishmania vector lives [1]. According to WHO, the number of leishmaniasis cases has been increasing during last two decades where more sensitivity with a high rate of Leishmania and AIDS co-infection cases is reported in patients suffering from AIDS in many areas [2-5]. Chemotherapy against Leishmania infestation, because of the toxicity and drug resistance, is not totally worthwhile [6, 7]. It has been shown that Th1 immune responses are crucial in immunity to Leishmania parasite [8] and because of the capability of single antigenic proteins in inducing Th1 immune responses, these proteins are now in main focus of vaccine candidates against the pathogen [9, 10].
Vaccination is postulated as the most appropriate opportunity for the prevention and safe treatment of all forms of the disease so developing a safe, effective and affordable anti-leishmanial vaccine is accounted as a promising approach for Leishmania researchers [11].
Several studies have been carried out to prove the immunogenicity of killed Leishmania parasite using different adjuvants such as Bacillus Calmette Guerin (BCG) [12] but no significant protection was observed following the injection of the vaccine. However, the mixture was safe and induced Leishmania Skin Test (LST) conversion with weak but measurable IFN-γ production. Using live Leishmania as a potent vaccine (leishmanization) has been considered as another strategy in the prevention of leishmaniasis [13]. Although, the live vaccine is of low cost and highly immunogenic, it still lacks standardization and quality control.
Genetically modified Leishmania is carried out either by mutagenesis and or gene targeting methods in which either a foreign gene is introduced into the parasite genome or by knocking out virulency genes [14]. Recently, recombinant proteins of Leishmania species have been used to produce the immunity against different species of Leishmania. Different proteins have been tested to be used as a vaccine including recombinant acylated surface protein B1 (HASPB1) and promastigote surface antigen-2 (PSA-2), which are immunogenic and can develop variable levels of immunity in different animal models.
Leishmania gp63 or leishmanolysin, an immunogenic Leishmania protein, is shown to be present in all Leishmania species and accounted as a good vaccine candidate in Leishmania studies [15, 16]. Injection of gp63 in saline, Complete Freud’s Adjutant (CFA), Bacillus Calmette Guerin (BCG) and Corynebacterium parvum induced significant protection in mice. So, cloning Leishmania gp63 into a BALB/ mouse cell line would be a feasible strategy in developing a potent cell line expressing an immunogenic Leishmania protein to be used for Leishmania vaccines.
2. Objectives
The main objective of this study was to clone and detect the gene encoding L. major gp63 in mouse CT26 cell line.
3. Materials and Methods
In this observational study, N-methylurethane-induced BALB/c murine colon carcinoma (CT-26) cells were gifted by Prof Ian Hart (St Thomas hospital) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) + 10% Fetal Calf Serum (FCS).
3.1. Preparation of L. major cDNA
CMV promoter pNUT L. major gp63 DNA was kindly gifted by Prof Mack Master (Department of Medical Genetics, University of British Columbia, Vancouver, Canada). The construct was first sequenced by MWG-Biotech using the primers ATCACGAAGTCGGTGTTG, GCACATCACCGAGGGCTT and AGAGCGAGGACGCCATCC and checked for mismatches against the sequence of the gene (code number Y00647) in the gene bank.
3.2. PCR Amplification
PCR was performed using a DNA thermal cycler (Thermo Hybaid, USA) for amplification of the whole gene. Primers were supplied by Sigma Genosys (UK). For amplification by PCR, 1 μL of cDNA was mixed with 5 μL of 10 × PCR buffer, 0.8 μL each of 10 mmol dNTP, 20 pmol each of primer solutions, 1.25 unit of thermostable Taq polymerase (Bioline), 1.5 mmol MgCl2 (Bioline) and then water was added to a total volume of 50 μL. The PCR program was set up as an initial melting step at 95ºC for 10 minutes followed by 35 cycles including 1 minute at 95ºC for denaturation, 45 seconds at 58ºC for annealing and 2 minutes or 45 seconds (only for screening) at 72ºC for extension and a final extension step at 72ºC for 10 minutes. PCR products were visualized using a 1.5% (w/v) agarose gel containing 1 μg/mL of ethidium bromide (BDH Laboratories, UK).
3.3. Ligation of L. major gp63 Gene and pcDNA3 Vector
The L. major gp63 gene produced by PCR using the primers containing HindIII and EcoRI restriction sites and the pcDNA3 vector were digested with relevant restriction enzymes and run onto the gel agarose. The gene and the vector were then cut and extracted from the gel by DNA extraction kit (Gene Flow) with accordance to the manufacturer’s protocol and ligated together using ligation enzyme (promega). The ligation was set up by adding 0.5 µL ligation enzyme, 1 µL buffer and 6.5 µL water to 2 µL of each DNA (adjusted at 10 µL). The ligated DNA was incubated at 4ºC overnight. The pcDNA3 vector containing L. major gp63 gene was reproduced by transforming E. coli and extracting the DNA from the bacteria.
3.4. Antibiotic Sensitivity Assay
In presence of a serial concentration of Geneticin (G418) (Sigma) from 50 to 900 µg/mL, 1×106/well CT26 tumour cells were cultured in 24-well plates; wells for each concentration of the antibiotic were put in duplicate. The cells were incubated at 37ºC with 0.5 CO2 for 10 days. The lower concentration (500 µg/mL) of the antibiotic in which all the CT26 tumour cell died in 7 - 10 days was chosen for transfection.
3.5. Transfection of CT26 Tumour Cells With L. major gp63
Lipofectamine 2000 (Invitrogen) was used to transfect CT26 tumour cells with pcDNA3 L. major gp63. Briefly, the culture of CT26 tumour cells at 1×106 per well in 24-well plates with 90% confluence was chosen. pcDNA3 L. major gp63 and lipofectamine 2000 were diluted in serum free DMEM media at 0.8 µg/50 µL and 2 µg/50 µL respectively and incubated for 5 minutes at room temperature. The diluted DNA and lipofectamine 2000 were mixed together and incubated again at room temperature for 20 - 30 minutes. After removing the supernatant, the DNA-lipofectamine was added to the CT26 cell culture and the culture was incubated at 37ºC with 0.5% CO2 for 4 - 6 hours. One milliliter per well DMEM media containing 10% FCS was then added to the culture. Sixteen until 24 hours later, the media was replaced with a fresh media complemented with 10% FCS and 500 µg/mL G418.
3.6. Screening the Expression of L. major gp63 Gene by RT-PCR
The stable transfected CT26 tumour cells were screened for the presence of the L. major gp63 gene by RT-PCR. Total RNA was isolated from the cells using RNA STAT-60 (AMS Biotechnology, UK) following manufacturer’s instructions. Briefly, CT26 L. major gp63 were cultured in T25 tissue culture flasks. After removing the media, 1 mL of RNA-STAT60 was added to the cell culture followed by incubating at room temperature for 5 minutes, adding 0.2 mL of chloroform, vigorously shaking for 60 seconds and leaving at room temperature for 3 minutes. The homogenate was then centrifuged at 14,000 rpm for 10 minutes and the aqueous phase was separated. Total of 0.5 mL of isopropanol was added the aqueous phase of samples followed by 8 minute incubation at room temperature and 15 minute centrifugation at 14,000 rpm. After removing the supernatant, the RNA pellet was washed with 75% ethanol, dried and resuspended in molecular grade water.
3.7. RNA was Then Reverse Transcribed into cDNA as Follow
Two microgram of RNA and 0.5 μg of oligo (dT15) primer (Biorad) were heated at 70ºC for 5 minutes and placed on ice. The following mixture was then added to the tube: Five microliter of 5 × reaction buffer, 1 μL of dNTPs (12.5 mmol) (Biorad), 25 units rRNasin Ribonuclease Inhibitor (Biorad), 200 units of M-MLV Reverse Transcriptase (Biorad), nuclease free water was added to make the final volume to 25 μL. Contents of the tube were gently mixed and heated at 39.2ºC for 80 minute followed by cooling on ice and heating at 95ºC for 5 minute and then storing them at 20ºC. PCR amplification was applied to screen the presence of L. major gp63 gene using the reverse transcribed RNA as template.
4. Results
4.1. L. major gp63 DNA Sequencing
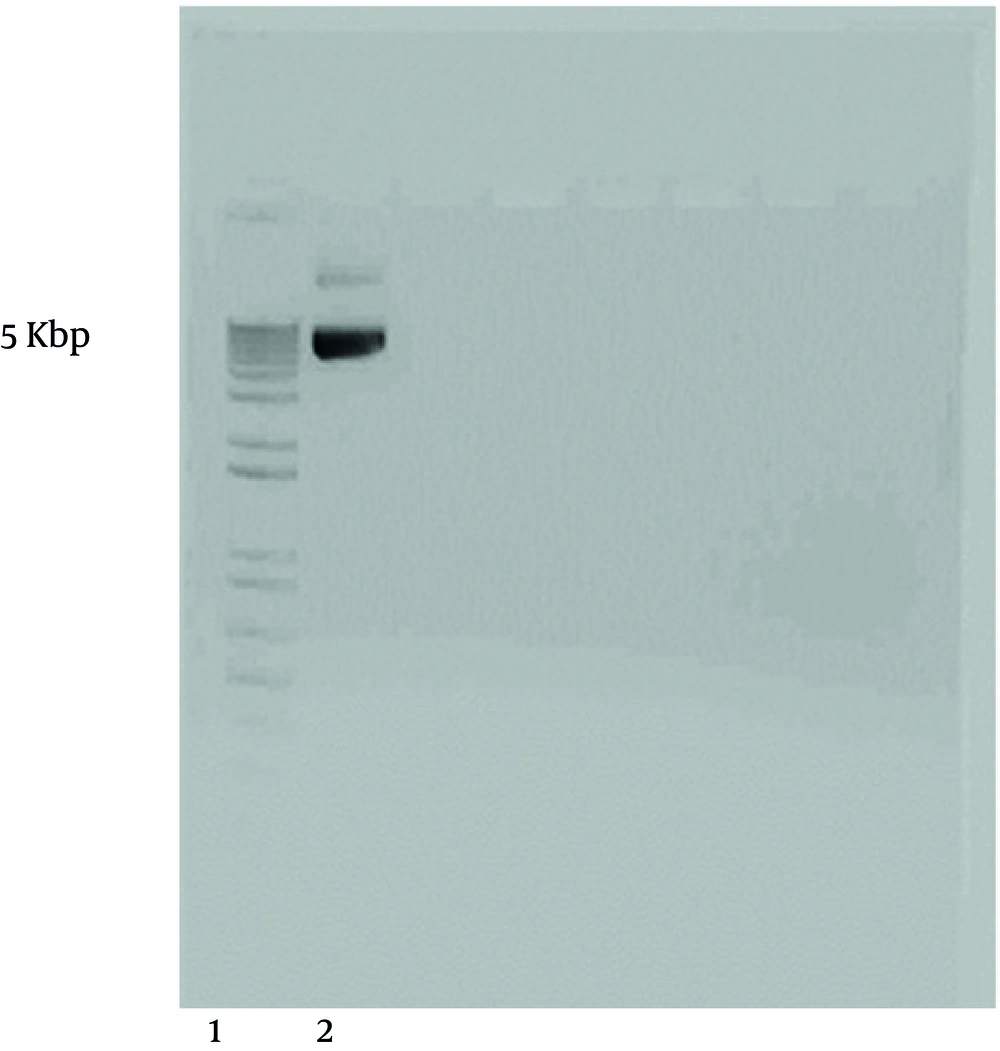
The sequence of the gene was evaluated (Figure 1) using specific primers and aligned with the sequence of the gene in the gene bank (see materials and methods). CMV promoter pNUT L. major gp63 DNA (Figure 2) was first bulked up by transformation of E. coli.
4.2. L. major gp63 Gene Sub-Cloning
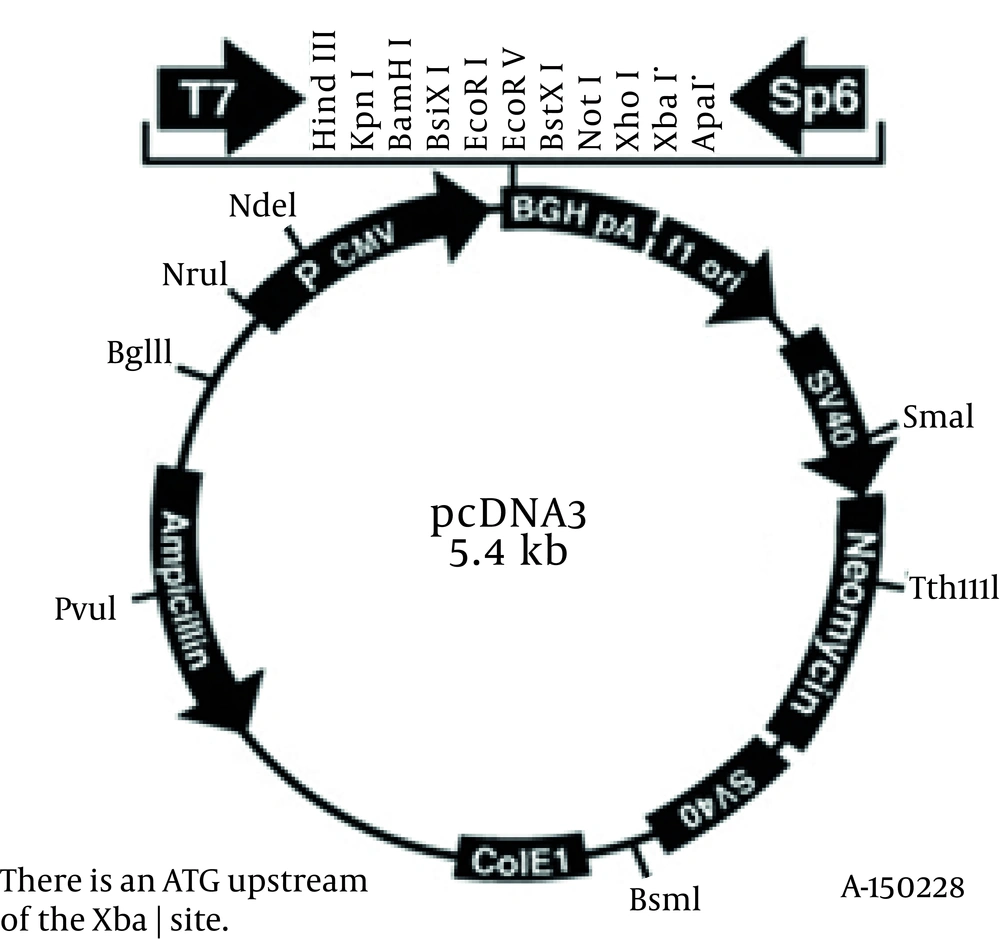
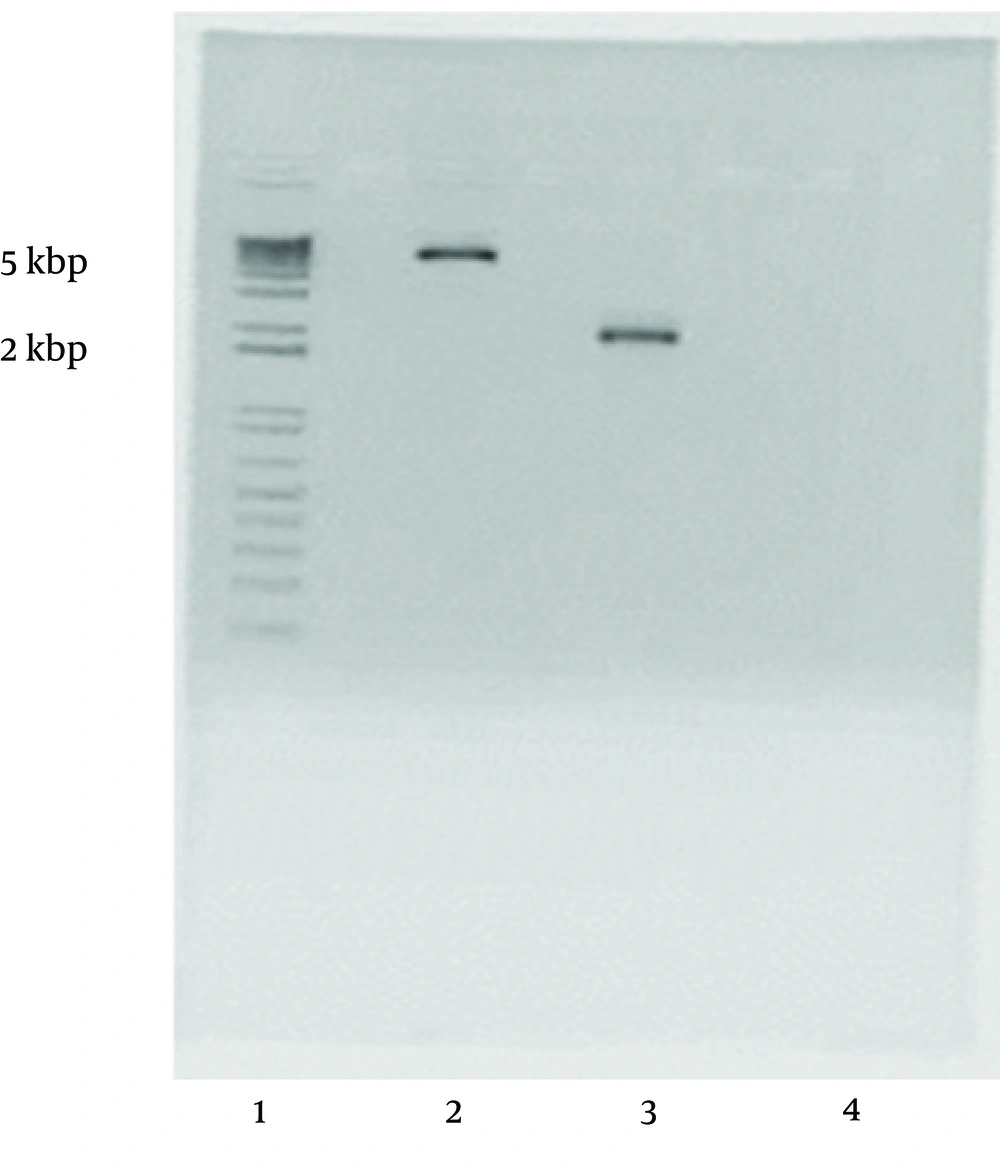
L. major gp63 gene was copied by PCR using two specific primers (5'- TCTAAGCTTCACCCGATCCATG TGCCG-3’ and 5'- AGAATTCCTGCACACTGGCGG CCGTTA -3') in which two restriction sites HindIII and EcoRI were engendered. The copied gene and pcDNA3 vector (Figure 3) were both cut by the relevant enzymes and the digested products were run onto the gel agarose (Figure 4). L. major gp63 and pcDNA3 bands were cut off the gel and the DNAs were extracted. The enzyme digested pcDNA3 vector and the L. major gp63 DNA were ligated together (see materials and methods). After ligation the existence of the gene in pcDNA3 vector were checked and the gene was completely sequenced and aligned with that of pNUT vector. No mismatches were found between the two sequences.
4.3. Transfection of CT26 Tumour Cells With L. major gp63
pcDNA3 L. major gp63 was applied onto CT26 tumour cells for transfection of the cells using lipofectamine 2000 (see materials and methods). The presence of the gene was examined with detection of its mRNA using RT-PCR. The following mouse GAPDH primers 5’-ACTCCACTCACGGCAAATTC-3’ and 5’-CCTTCCA CAATGCCAAAGTT-3’ were used to check the efficacy of the mRNA extracted from the CT26 transfected cells and specific L. major gp63 primers used to check the presence of the gene (Figure 5).
5. Discussion
The results of our study clearly showed that Leishmania gp63 successfully transfected into CT26 mouse cell line and these cells have a high potency in accepting Leishmania genes and expressing the parasitic proteins.
Developing specific resistance after infection approves the feasibility of vaccine approaches against Leishmania [17]. Different attempts have been made to generate Leishmania vaccines using recombinant immunogenic Leishmania proteins expressed in microbial vectors but none has yet been introduced as a vaccine for Leishmania [18, 19]. However, the potency of mammalian cells in expression of Leishmania immunogenic genes is not yet fully cleared.
Cloning and characterization of antigenic Leishmania genes is now postulated as a promising approach in Leishmania vaccination. So far, different studies have been conducted to clone, express or manipulated Leishmania genes. Cloning and characterization of a gene encoding class I nuclease in L. infantum has already been carried out by Farajnia et al. [20]. Also, there are other reports on cloning and expression of different Leishmania genes such as HSP70 and glucose-regulated protein 94 (GRP94) gene from L. infantum [21, 22], surface protein encoding genes from L. major [23, 24] and L. donovani avastin gene [25]. Some other researchers tried to find similarity between genes cloned in other protozoa and the Leishmania ones in order to develop immunity against Leishmania [26]. There are some attempts for establishing cell lines specific for Leishmania, which were promising [17]. One of the characterised immunogenic proteins in all Leishmania species is Leishmania gp63 or leishmanolysin [16]. It has been shown that vaccination based on gp63 DNA or recombinant gp63 protein is potential to induce T cell stimulation against Leishmania in human producing high levels of INF-γ from effector T cells [27, 28]. Our previous studies also proved the immunogenicity of peptides predicted for either class I or II derived from this protein [29, 30]. We also showed that expression of Leishmania gp63 protein in host's cells by either injection or using the gene gun can elicit immunity to Leishmania [31]. Similar results have been demonstrated for LACK antigen where in presence of recombinant IL-12 evokes CD4+ T cell inducing protection in mice against L. major infection [32]. Therefore, cloning Leishmania gp63 into a BALB/ mouse cell line would be a feasible strategy in developing a potent cell line expressing an immunogenic Leishmania protein to be used for Leishmania vaccines.
In present study, it was tried to clone a gene derived from protozoan cells and express it in mammalian cells in order to develop a potent mammalian cell line expressing an immunogenic Leishmania protein, which can be used for Leishmania vaccines. To clone the gene, L. major gp63 DNA was first cut from a week construct (pNUT vector) and sub-cloned into pcDNA3 vector using specific primers.
In conclusion, a full length of L. major gp63 gene was cloned into the mouse CT26 cell line and the stable expression of the gene was evaluated. The results confirmed that Leishmania genes can be successfully expressed in mammalian cell lines, which could be used as a potential vaccine against Leishmania.