1. Background
Coxiella burnetii is an obligate intracellular bacterium that causes the zoonotic disease Q fever with a worldwide distribution. It has a wide range of hosts including mammals such as ruminants, dogs, cats, non-mammal primates, wild rodents, small mammals, big game, and non-mammals such as reptiles, amphibians, birds, fish and ticks [1, 2].
Cattle, sheep and goats are the main sources of infection in humans [3, 4]. Infected animals excrete C. burnetii into the environment via birth products such as the placenta. This bacterium is very stable in different environments. It is also highly infectious and one to ten organisms can cause Q fever in humans [5]. Also, C. burnetii can be present in milk, urine, feces, vaginal mucus and semen. In milk, it can be secreted for 8 days in ewes and up to 13 months in cattle [6]. The consumption of contaminated raw milk does not seen to represent an efficient route of disease transmission, however bulk milk samples is an important specimen for epidemiological survey on dairy herds [7]. In order to management, prevention, control and treatment of Q fever in animal and human, early and accurate detection of C. burnetii is very necessary. Previous studies on the prevalence of C. burnetii in dairy cows were based mainly on serologic tests that detect antibodies that could have been introduced months earlier [8]. Isolation of C. burnetii is very difficult and dangerous. Recently, PCR has been used to detect C. burnetii. PCR is a safe, sensitive and specific method for the detection of C. burnetii in different samples [9]. Several target genes that are used for specific C. burnetii identification include: the superoxide dismutase (Sod B) gene, com1 encoding a 27 kDa Outer membrane protein, the heat shock operon encoding two heat shock proteins (htpA and htpB), isocitrate dehydrogenase (icd), the macrophage infectivity potentiator protein (cbmip) and a transposon-like repetitive region of the C. burnetii genome (Trans) [10].
2. Objectives
Since the clinically healthy cattle are the main source of C. burnetii infection in Iran [11], in the present study, we evaluated the sensitivity of PCR with 3 different primers for detection of C. burnetii in bovine bulk milk samples.
3. Materials and Methods
In this cross-sectional study 70 bovine bulk milk samples were collected randomly from dairy herds in Jahrom city in the Southern of Iran in 2010. The samples were immediately transported to the laboratory and were tested. One militer of raw milk was centrifuged. This procedure was performed to isolate the bacterial cells in pellet of the milk samples. After removing the cream and milk layers [12], DNA was extracted from the pellet by a genomic DNA purification kit (Cinna Gen Co., Iran) according to the manufacturer’s protocol. DNA samples were stored at -20°C until they were used. In this study, we used 8 genomic primers targeting 3 different genes: A) Trans1 and Trans2 were designed based on the transposing-like repetitive region of the C. burnetii genome [13]. The length of the genome target for amplification was expected to be 687 bp. B) OMP1, OMP2, OMP3 and OMP4 were designed from the nucleotide sequence of the com1 gene encoding a 27 kDa outer Membrane Protein (OMP) as previously described [14]. The expected amplification product of the target sequence with OMP1, OMP2 was 501 bp long and with OMP3, OMP4 was 438 bp long. C) The new primers Coc-F and Coc-R were designed based on the 16S rRNA gene in the present study. The length of the predicted product was 242 bp.
All oligonucleotide primers were obtained from a commercial source (Cinna Gen Co., Iran). The sequence of the primers is shown in Table 1. The Trans-PCR thermal program was carried out according to the method described in [9]. The amplification was performed in a total volume of 25 μL containing 2 µL of DNA sample, 1.5 mM MgCl2 ,0.2 mM (each) dNTPs, 0.2 µM primer Trans1, 0.2 µM primer Trans2 and 1 U/reaction of Smar Taq DNA polymerase (Cinna Gen Co., Iran). The thermal program was carried out under the following conditions: five cycles of 94°C for 30 second, 66 - 61°C (the temperature was decreased by 1°C between consecutive steps) for 1 min, 72°C for 1 min and then 40 cycles of 94°C for 30 s, 61°C for 30 s and 72°C for 1 min. For the Nested PCR with primers OMP1 - OMP2 and OMP3 - OMP4, the first amplification was performed in a total volume of 25 μL, containing 2.5 µL of DNA sample, 1.5 mM MgCl2 ,0.2 mM (each) dNTPs, 1 µL primer OMP1, 1 µL primer OMP2, and 2.5 U/reaction of Smar Taq DNA polymerase (Cinna Gen Co., Iran). The PCR assay was done at 94°C for 4 min and then for 30 cycles of 94°C for 1 min, 56°C for 1 min, 72°C for 1 min and a final extension at 72°C for 5 min in a DNA thermal cycler. In the second amplification, the reaction mixture was the same as that in the first amplification, except for primers and DNA templates. In this amplification, primers OMP3 OMP4 were used and the first amplification product was used as the DNA template. The PCR assay was performed at 95°C for 4 min and then for 30 cycles of 94°C for 1 min, 57°C for 1 min, 72°C for 1 min and a final extension at 72°C for 5 min. For the semi nested PCR with primers Coc-F and Coc-R, the amplification was performed in a total volume of 25 μL containing 2 µL of DNA sample, 1.5 mM MgCl2, 0.2 mM (each) dNTPs, 0.2 µM primer Coc-F, 0.2 µM primer Coc-R and 1 U/reaction of Smar Taq DNA polymerase (Cinna Gen, Iran).
| Target Gene | Primers | Primers Sequence | Size (bp) |
|---|---|---|---|
| OMP1 | AGT AGA AGC ATC CCA AGC ATT G | 501 | |
| OMP2 | T GAA GCG CAA CAA GAA GAA CAC | 501 | |
| OMP3 | GC CTG CTA GCT GTA ACG ATT G | 438 | |
| OMP4 | TTG GAA GTT ATC ACG CAG TTG | 438 | |
| 687 | |||
| Trans1 | TAT GTA TCC ACC GTA GCC AGT C | ||
| Trans2 | CCC AAC AAC ACC TCC TTA TTC | ||
| 242 | |||
| Coc-f | GTA ATA TCC TTG GGC GTT GAC G | ||
| Coc-r | ATC TAC GCA TTT CAC CGC TAC AC |
The PCR Primers Used for the Detection of Coxiella burnetii Genes
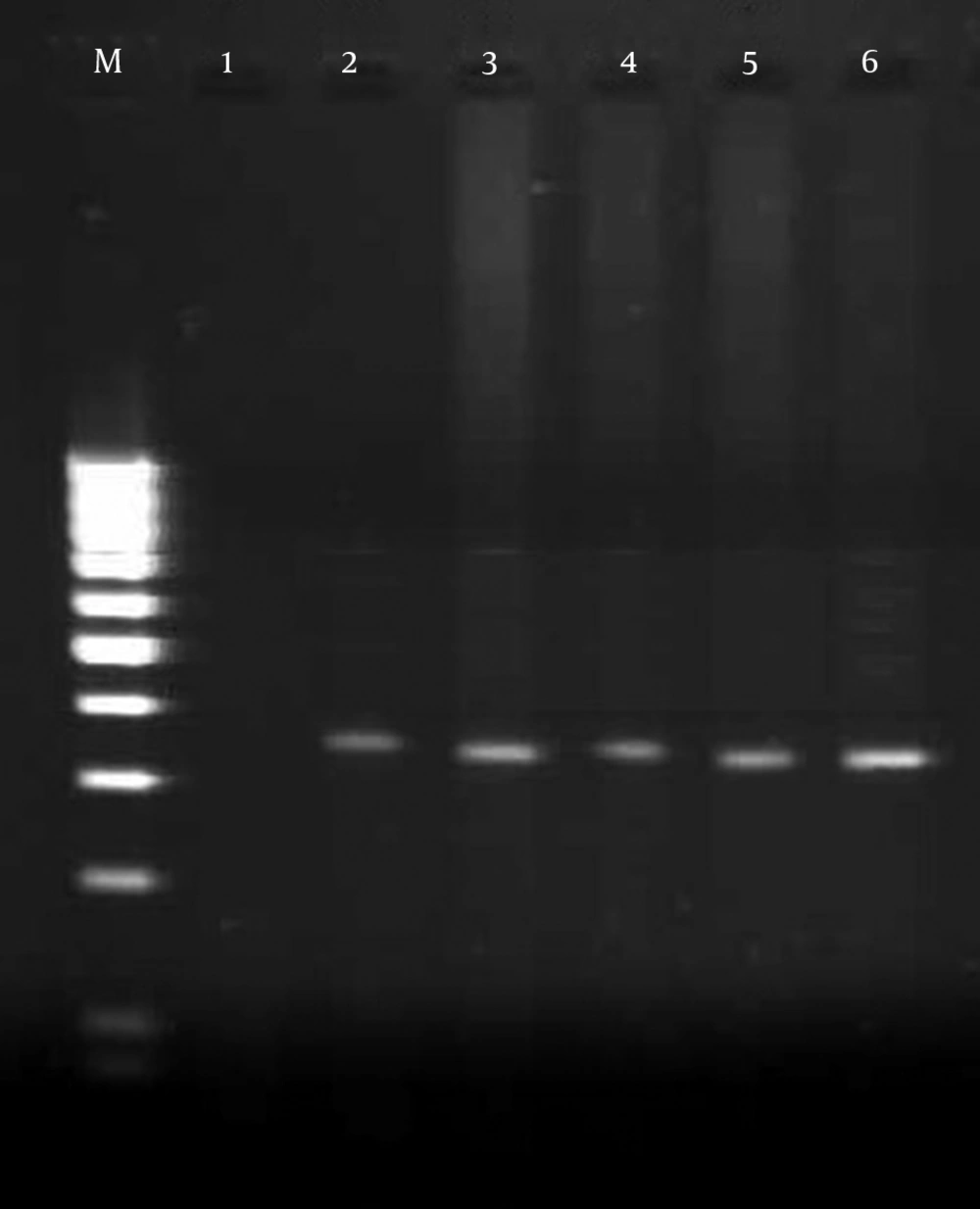
The thermal program was carried out under the following conditions: 95°C for 5 min and then for 32 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 1 min and a final extension at 72°C for 5 min. All of PCR reactions were performed in a DNA thermal cycler (Techne). In this study, we used positive and negative controls in each PCR run. C. burnetii DNA (serial Number: 3154; Genekam Biotechnology AG, Duisburg, Germany) was used as the positive control and negative controls were reaction mixtures without a DNA template. Sterile distilled water was used instead of a DNA template. DNA samples of C. burnetii and 5 other bacteria were used in the semi-nested PCR assay in order to evaluate the specificity of new primers Coc-F and Coc-R. The bacteria used in this test were, Staphylococcus aureus, Escherichia coli, Brucella abortus, Brucella melitensis, and Listeria monocytogenes. The PCR-amplification products (OMP1 - OMP2: 501 bp; OMP3 - OMP4: 438 bp; trans1 - trans2: 687; CocF - CocR: 242 bp) were examined by electrophoresis in a 1.5% agarose gel, visualized under UV and photographed by gel documentation (U GENIUS-SYSGENE).
4. Results
4.1. Specificity of Semi-Nested PCR
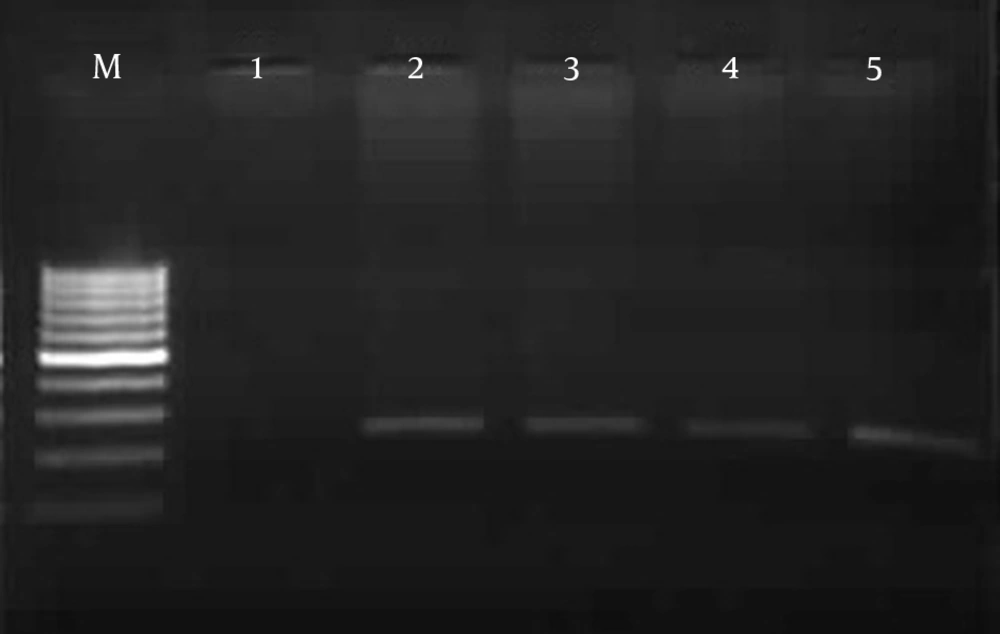
In the semi-nested PCR assay with new primers Coc-F and Coc-R only one specific band was observed with the expected size (242 bp) of C. burnetii.
4.2. PCR
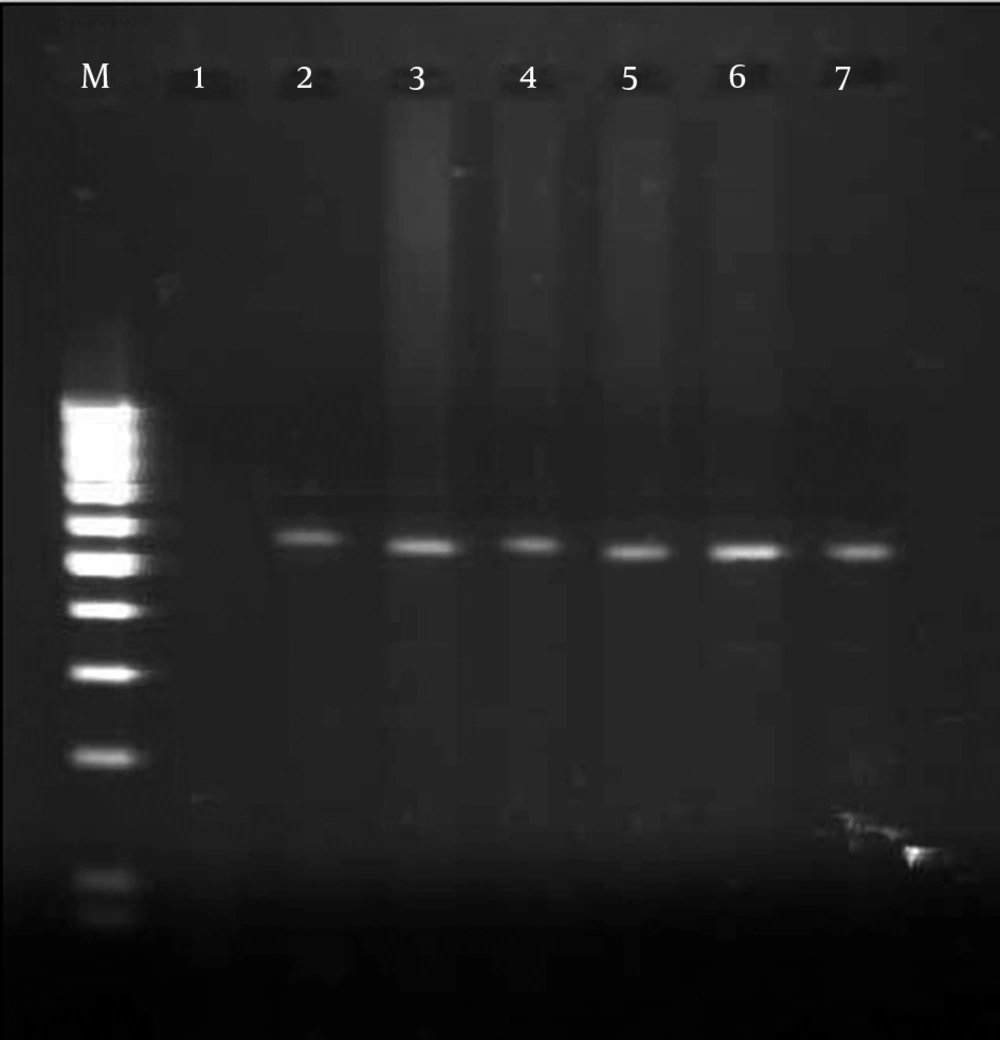
In total, 7 of 70 tested samples were positive with primers OMP1, OMP2, OMP3 and OMP4. The primers OMP1 - OMP2 and OMP3 - OMP4 amplified the predicted products of the 501 bp DNA in the first amplification and the 438 bp DNA in the second amplification of PCR (Figure 1). Also 10% of samples were positive with primers CocF - CocR and showed the 242 bp PCR product on agarose gel (Figure 2). While 12 positive samples were observed with primers Trans1 - Trans2, in amplification with these primers the bands appeared at approximately 687 bp, which was in line with the expected length for detection of C. burnetii (Figure 3). The results of the different methods are shown in Table 2.
| Methods | Numbers of Steps | Numbers of Positive Samples, % | Sensitivity | Specificity |
|---|---|---|---|---|
| 1 | 12 (17.14) | More than other assays | - | |
| 2 | 7 (10) | - | - | |
| 2 | 7 (10) | - | Acceptable |
Sensitivity and Specificity of the Different Methods for Detection of C. burnetti
5. Discussion
In this study we evaluated the sensitivity of PCR with 3 different primers for detection of C. burnetii in bovine bulk milk samples. The Trans-PCR showed more positive samples (17.14%) while the frequency of C. burnetii by OMP-PCR and Coc-PCR in the tested samples was 10%. This suggests that PCR with primers Trans1 and Trans2 are highly sensitive and useful for the detection of C. burnetii.
Previous studies on the prevalence of C. burnetii in dairy bovine were based mainly on serologic tests that detect antibodies that could have been introduced months earlier [8]. Also C. burnetii is classified as a select agent and a CDC (Centers for Disease Control and Prevention) category B bioterrorism agent, with current research on the agent requiring specialized high-containment biosafety level-3 facilities [15]. Recently, PCR have been used to detection of C. burnetii. PCR is usually used for the diagnosis of Q fever in ruminants in research and clinical works [9]. Although the prevalence of C. burnetii is likely to be low in milk, PCR can be used for the detection of C. burnetii in this matrix [13]. Several target genes are used for detection of C. burnetii by PCR. Most of studies have focused on detection and determination of the rate of C. burnetii by PCR, while a little information is available about the comparison of different primers.
The purpose of the present study was to evaluate the sensitivity of PCR with 8 different primers targeting 3 genes for the detection of C. burnetii in bulk milk samples. These primers were designed based on different target genes. Com1 and 16S rRNA are single copy genes while IS1111 is present at multiple copy numbers (7 to 110 copies), depending on the strains of C. burnetii [16]. So it was expected that some of the Trans-positive samples would be negative using the com1 and 16S rRNA assay.
In this test 7 out of 70 milk samples were positive for C. burnetii targeting com1 and 16S rRNA. The number of positive samples with Trans-PCR was larger than by any other assay. The use of Trans-primers for the amplification of IS1111 allows the sensitivity of the assay to be increased and this is because of the presence of several copies in the Coxiella genome. Also, Trans-PCR was run in one step whilst nested PCR and semi-nested PCR with other primers were run in two steps. Run of PCR in two steps is time consuming and will increase risk of contamination between two steps. Due to running the PCR in one step and the larger number of positive samples, the Trans-PCR can be more sensitive, reliable, and an easier and faster method for the detection of C. burnetii in bulk milk samples. These results are similar to a study in France, which Berri et al. showed Trans-PCR to be very highly specific and sensitive for the direct detection of C. burnetii in genital swabs, milk and fecal samples from ewes. Also they pointed that the high degree of efficacy of the trans-PCR can be attributed to the fact that the targeted region exists in at least 19 copies in the C. burnetii Nine Mile, phase I, genome, which gives the trans-PCR a level of sensitivity 100 times higher than that of the PCR assay [9]. In a similar study, Kim et al. in the USA showed the higher sensitivity of Trans-PCR in detection of C. burnetii. Also they reported that the trans-PCR assay detects C. burnetii in samples immediately, unlike serologic assays that detect antibodies that could have been introduced months earlier [8]. Vaidya et al. reported that the PCR assay with primers targeting IS1111, the repetitive, transposon-like element (Trans-PCR), is very specific and sensitive for the detection of C. burnetii in clinical samples [17]. In this research the Trans-PCR showed 17.14% positive samples while the frequency of C. burnetii by OMP-PCR and Coc-PCR in the tested samples was 10%. Since bulk milk samples is an important specimen for epidemiological survey on dairy herds, we evaluated the sensitivity of PCR with 3 different primers for detection of C. burnetii in bovine bulk milk samples. The results of this study suggest that the IS1111 assay is reliably detecting C. burnetii genomic DNA in milk samples and PCR with primers Trans1 and Trans2 are highly sensitive and useful for the detection of C. burnetii.
The results of this study are limited to the PCR-based methods for detection of C. burnetii in the bulk milk samples, so we cannot compare the specificity and sensitivity of PCR with other methods such as ELISA. This study suggests that in order to obtain reliable results, the large numbers of samples should be analyzed in subsequent studies. Also it is better to compare different methods for detection of C. burnetii.