1. Background
Crimean Congo hemorrhagic fever virus (CCHFV), an extremely virulent pathogen, belongs to the family Bunyaviridae, with ticks serving as its primary reservoir (1). The genome of CCHFV consists of three segments (S, M, and L), which encode viral proteins (2). The CCHFV is a tick-borne RNA virus with a widespread distribution across Africa, the Middle East, Asia, Southeastern Europe, and Spain (3). The geographic dissemination of this disease is the most extensive among tick-borne infections. Iran is one of the endemic countries, with human cases reported in nearly all regions (4). Early detection is essential for preventing further spread of the infection (5). While the disease is asymptomatic in infected animals, it can cause severe illness in humans (6). The incubation period is typically 3 - 7 days, with sudden onset of myalgia, headache, and fever, which can progress to a severe hemorrhagic syndrome (7). The recurrence of CCHFV poses a serious public health threat, as it is highly contagious, often fatal, capable of causing nosocomial infection, and difficult to treat, prevent, and control (8).
Viral isolation is the gold standard for CCHFV diagnosis but must be conducted in biosafety level 4 facilities. Furthermore, viral isolation is not error-free, and cell cultures have low sensitivity, typically detecting only high viremia levels during the first five days of disease (9). Recently, molecular-based techniques have been preferred for viral genome detection, as virus isolation requires high biosafety laboratories. Additionally, serological methods may yield false negatives during the early acute phase of CCHFV infection (10, 11). Nested polymerase chain reaction (PCR) is one molecular method used for CCHFV detection; however, it is time-consuming, and cross-contamination has been reported (12). Real-time PCR has been employed to address these issues, but it is expensive. Recently, the loop-mediated isothermal amplification (LAMP) assay has emerged as a novel gene amplification technique, offering high accuracy for identifying specific regions of target DNA (13). The reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay, performed under isothermal conditions at 63°C, requires no specialized equipment (5). It utilizes 4 - 6 primers and completes in under one hour, making it a simple and effective tool for rapid molecular detection of various bacteria, viruses, fungi, and parasites (13, 14). The LAMP method is highly sensitive, capable of detecting and amplifying fewer than 500 copies of DNA (13, 15). Because LAMP recognizes six regions of the target sequence, its results are expected to be more specific than those of PCR (16, 17).
2. Objectives
Given the need for rapid, accurate, and convenient CCHFV detection in areas with limited laboratory resources, the present study aimed to develop a RT-LAMP assay for the rapid detection of the small (S) RNA segment of CCHFV. The RT-LAMP primers were designed based on a highly conserved region of the S segment of the viral genome.
3. Methods
3.1. Sequence Alignments
The S segment sequences of CCHFV were retrieved from GenBank at the National Center for Biotechnology Information (accession numbers: AY366376, AY366373, AY366379, AY366378, AY366375, AY366377, AY366374, GU456725, GU456728, GU456724, GU456727, GU456726, AY905654, AY905653, JF798869, JF798867, JF798866, AY905655, GU456723, JF798868). Sequence alignments were performed using CLC Sequence Viewer 7.5 (CLC bio, Aarhus, Denmark).
3.2. Loop-Mediated Isothermal Amplification Primer Design
Six specific primers for the LAMP assay, including loop primers, were designed from a conserved region of the S segment using the online tool Primer Explorer V4. Primer specificity was verified using the National Center for Biotechnology Information’s Basic Local Alignment Search Tool (BLAST), and the primers (Table 1) were synthesized by Bioneer (Daejeon, Korea).
| Primers | Sequence (5´-3´) | Reference |
|---|---|---|
| F3-CC | CCTATATACGAATGTGCCTGG | This study |
| B3-CC | TTGACACGGAAGCCTATG | |
| FIP-CC | CCATGATTTAATGGTTCCTGCATTT-TAGCTCCACTGGCATTGT | |
| BIP-CC | AGTTATACTGAGCTGAAAGTTGAGG-TCTTTCCTCCACTTGAGAG | |
| LF-CC | AACCACTCCAGTCCCTTCTTA | |
| LB-CC | TCCCAAAATAGAACAGCTTTCCAAC |
3.3. Sample Preparation
Tick specimens contaminated with CCHFV were collected in collaboration with the Veterinary Center of Sistan and Baluchistan province and transferred to the Parasitological Laboratory of the University of Tehran. After genus and species identification, the ticks were sent to the Biotechnology Research Center of AJA University.
3.4. RNA Extraction, Complementary DNA Synthesis, and Polymerase Chain Reaction
RNA was extracted using a commercial RNA extraction kit (Qiagen, Valencia, CA), and complementary DNA (cDNA) was synthesized using the RevertAid First Strand cDNA Synthesis kit (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA, cat# K1621). The PCR amplification of the target gene using F3CC and B3CC primers was performed in a 25 µL reaction volume [12.5 µL 2X Reaction Mix, 1.5 µL F3 (10 pmol/L), 1.5 µL B3 (10 pmol/L), 1 µL Taq Polymerase 1 U/µL, 5 µL cDNA, and 4.5 µL distilled water]. A negative control reaction was conducted without template addition. The PCR products were confirmed by electrophoresis on a 2% agarose gel.
3.5. Preparation of Standard Plasmid Using TA Cloning
After PCR amplification of the target gene with F3CC and B3CC primers, the product was purified using a PCR Purification Kit (Bioneer, Korea), yielding a 200 bp fragment. This fragment was ligated into the pTZ57R/T vector using T4 DNA ligase according to the InsTAclone PCR cloning kit instructions (Fermentas, USA). The ligated product was transformed into Escherichia coli Top10 F’ competent cells, which were plated on Luria-Bertani medium (Merck, Germany) containing ampicillin (50 mg/mL) and tetracycline (30 mg/mL) and incubated at 37°C for 24 hours. Recombinant clones were identified via blue/white screening, and several white colonies were selected for further analysis. Plasmids were extracted using a BS413 EZ-10 Spin Column Plasmid DNA Kit (Bio Basic, Canada). Insertion of the target gene was confirmed by a second round of PCR with F3CC and B3CC primers and by cycle sequencing. The verified plasmid, designated “pTZ-CC”, was used as a positive control in the LAMP assay.
3.6. Loop-Mediated Isothermal Amplification Assay
The LAMP reaction was performed in a 25 µL volume containing 12.5 µL LAMP reaction mix [1.4 mM each deoxynucleotide triphosphate, 1 M betaine (Sigma-Aldrich, USA), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH2)SO4, 8 mM MgSO4, 0.1% Triton X-100], 1 µL fluorescent detection reagent [625 μM calcein (Dojindo Molecular Technologies, Inc, Japan) and 12.5 mM MnCl2 (Sigma-Aldrich, USA)], 1.5 µL LAMP primer mix (FIP-CC and BIP-CC: 40 pmol/µL, F3-CC and B3-CC: 5 pmol/µL, LF-CC and LB-CC: 20 pmol/µL; Table 1), 8 µL distilled water, 1 U Bst DNA polymerase large (L) fragment (1 µL) (New England Biolabs, Ipswich, MA, USA), and 2 µL pTZ-CC plasmid (280 ng/mL). To observe turbidity in the LAMP experiment, additional tubes were prepared under identical conditions without the fluorescent detection reagent. Negative controls contained all components except template DNA. Reactions were incubated at 65°C for 60 minutes in a Loopamp real-time turbidimeter (LA-320C; Teramecs, Japan) and then terminated at 80°C for 5 minutes to inactivate Bst DNA polymerase. Products were analyzed by real-time turbidity curves, 2% agarose gel electrophoresis, and visual inspection for fluorescence or turbidity. Final validation was performed via cycle sequencing with the F3-CC and B3-CC primers, with results confirmed by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
3.7. Loop-Mediated Isothermal Amplification on Contaminated Ticks
Ticks were collected in rural areas of West Azerbaijan province in March and April 2017. A total of 60 ticks were gathered from various body parts of ruminants (sheep, goats, and cows) and transferred to the laboratory in test tubes. Identification was based on morphological features and systematic keys. Each tick was washed twice with phosphate-buffered saline (PBS, pH 7.4) and crushed in 300 µL PBS using a mortar and pestle. Total RNA was extracted using the Qiagen RNA extraction kit and cDNA synthesized using the RevertAid First Strand cDNA Synthesis kit (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA, cat# K1621). The RT-PCR was used to assess sample contamination with CCHFV (18). The cDNA from all RT-PCR-positive samples and five negative samples was evaluated with the S-LAMP method as described above.
3.8. Evaluation of the Loop-Mediated Isothermal Amplification Method with Artificially Contaminated Samples
To assess the performance of the S-LAMP method in detecting the S segment of CCHFV in blood, artificially contaminated samples were prepared. Ten microtubes, each containing 200 µL of blood from healthy volunteers, were spiked with pTZ-CC plasmid to a final concentration of 1 µg/200 µL. Five microtubes containing 200 µL of blood were used as negative controls. Samples were incubated for 96 hours at 4°C, then DNA was extracted with the Genomic DNA kit (Bio Basic Inc., Canada), and S-LAMP was performed on all samples.
3.9. Sensitivity Assay of the Loop-Mediated Isothermal Amplification Method
To determine the limit of detection (LOD) of the S-LAMP method, a serial dilution of the target gene was prepared. The pTZ-CC plasmid (620 ng/mL) was serially diluted 10-3 to 10-10. The LAMP reaction was performed at 65°C for 60 minutes for each dilution. Results were analyzed using amplification curves, direct turbidity assay, and electrophoresis. In each detection method, the final dilution showing a specific amplification signal was considered the LOD.
4. Results
4.1. Polymerase Chain Reaction with F3-CC and B3-CC Primers
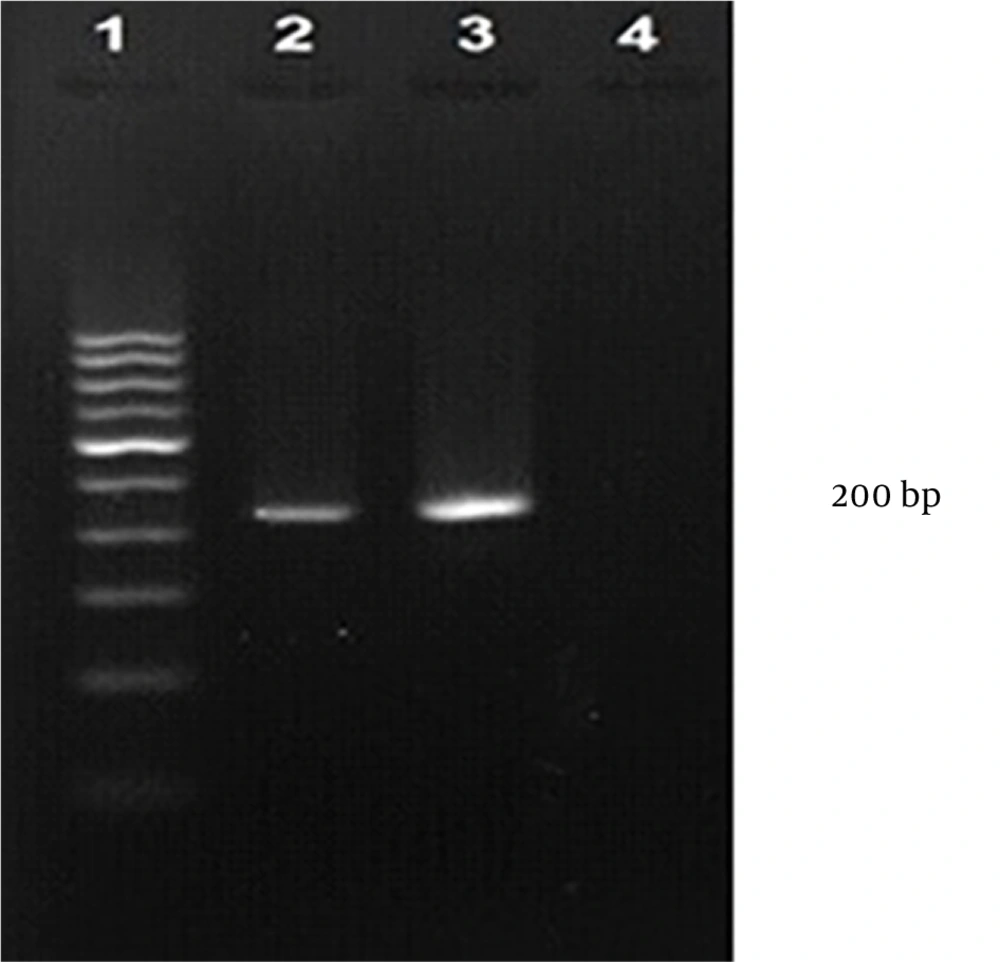
The consensus sequence of the S gene was used for primer design with Primer Explorer V4 (http://primerexplorer.jp/e/), and in silico specificity was confirmed using BLAST and Primer-BLAST (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/). Analysis with CLC Sequence Viewer 7.5 (CLC bio, Aarhus, Denmark) indicated that the designed primers could amplify a 200 bp fragment of the target gene. After RNA extraction and cDNA synthesis, amplification of the S segment with F3-CC and B3-CC primers produced a clear 200 bp band on a 2% agarose gel (Figure 1).
4.2. Cloning of the Polymerase Chain Reaction Product for Positive Control
To obtain a stable positive control, the PCR product was cloned into a vector. After colony screening and PCR, a positive clone was selected and named pTZ-CC. Sequencing confirmed insertion of the S sequence, and the plasmid was stored at -20°C for future use (Figure 2).
4.3. Analysis of Loop-Mediated Isothermal Amplification Products
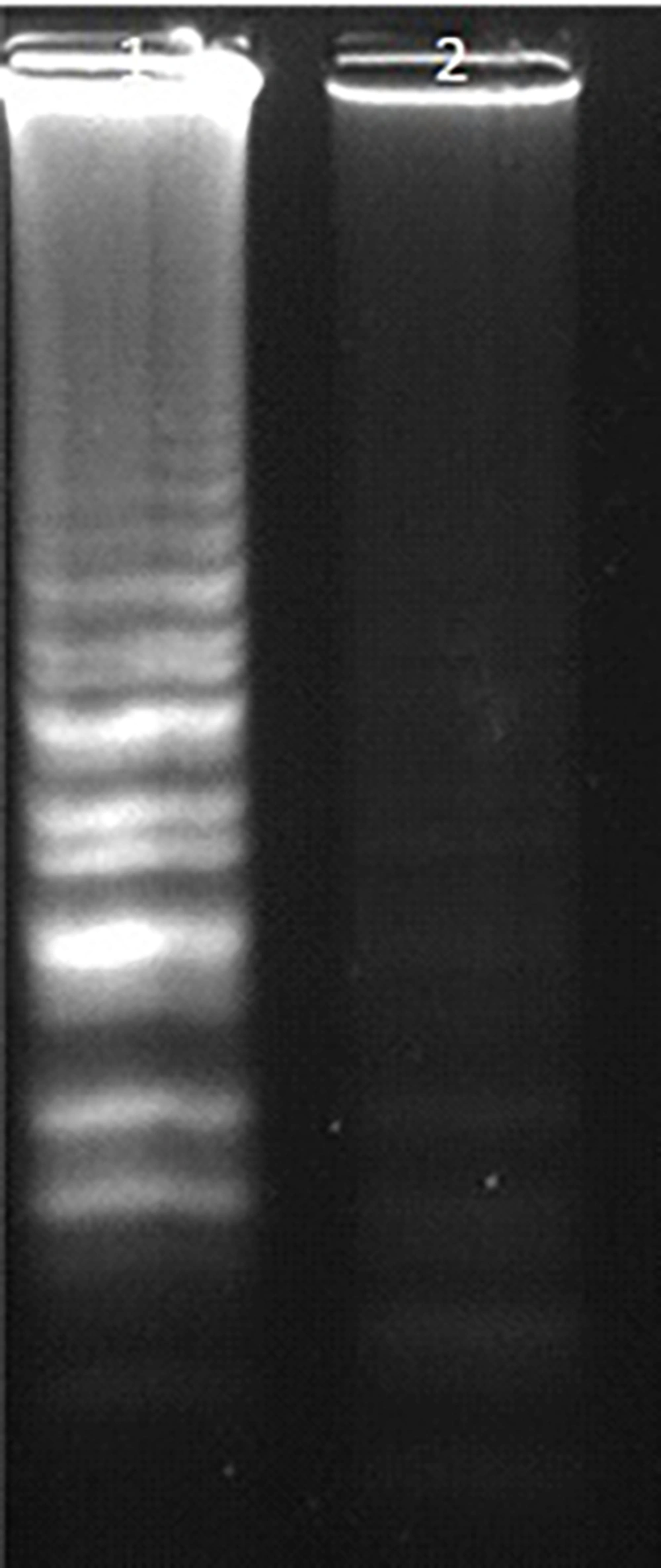
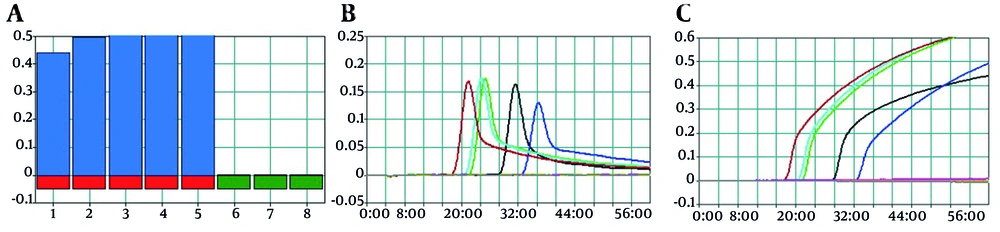
In tubes with successful isothermal amplification, turbidity due to white magnesium pyrophosphate precipitation was visible to the naked eye. Under ultraviolet light, positive tubes exhibited green fluorescence resulting from the interaction of calcein with magnesium ions. Judgment curves generated by the LA-320C real-time turbidimeter confirmed amplification of the S sequence (Figure 3). Electrophoresis showed a strong ladder-like DNA amplification pattern (Figure 4). The BLAST analysis of the products confirmed their identity.
4.4. Sensitivity of the Loop-Mediated Isothermal Amplification Method
The lowest dilution of pTZ-CC plasmid yielding significant amplification in the S-LAMP assay was 10-7 (62 fg). Copy number calculation using an online tool (technologynetworks) yielded 1.8 × 104 copies. Sensitivity was confirmed via judgment and amplification curves from the LA-320C turbidimeter (Figure 5). Electrophoresis of S-LAMP reactions with different pTZ-CC dilutions showed that the minimum detectable concentration was 62 fg/µL (Figure 6).
4.5. Evaluation of the Loop-Mediated Isothermal Amplification Method in Contaminated Ticks
After S-LAMP amplification on contaminated ticks, tubes were exposed to ultraviolet light. The fluorescent signal was observed in all tubes previously positive by RT-PCR, as well as in the positive control tube (Figure 7). All negative samples remained negative in this assay.
4.6. Evaluation of the Loop-Mediated Isothermal Amplification Method Using Artificially Contaminated Samples
All 10 artificially infected samples were positive by S-LAMP, while all five uninfected control samples tested negative.
5. Discussion
This study utilized the LAMP method to detect the CCHFV genome in contaminated ticks. Sensitivity assessment was performed using serial dilutions of the pTZ-CC plasmid and specific primers at a single temperature. The LAMP method is faster, less expensive, and can be completed in a shorter time than real-time PCR, requiring only a simple thermal block at 65°C, with detection possible by directly observing the resulting turbidity. This method is suitable for rapid, accurate, and affordable molecular diagnostics, with broad applicability in diagnostic laboratories, particularly in resource-limited and mobile laboratory settings (5). The LAMP assay is also suitable for molecular detection of pathogens in treated blood samples, even in health care locations in developing countries (19).
In a study by Ito et al., LAMP assays developed for influenza A (HA1, HA3) and B viruses demonstrated superior sensitivity and accuracy for rapid detection compared to commercial immunochromatography tests (20). Imai et al. established an H5-RT-LAMP system for the rapid detection of avian H5 influenza, showing their primers could detect H5N1 influenza virus with 100-fold greater sensitivity than RT-PCR (21). Boehme et al. applied the LAMP technique in microscopy laboratories in developing countries for tuberculosis diagnosis, demonstrating its feasibility with limited clinical equipment (22). Moslemi et al. compared LAMP and PCR for hepatitis B virus detection; among 104 specimens, 95 (91.34%) were PCR-positive, while 101 (97.11%) were positive by LAMP. Of nine PCR-negative samples, six were positive by LAMP, indicating higher sensitivity and specificity (23).
The CCHFV genome is a single-stranded, negative-sense RNA comprising three segments: The L fragment (6.5 - 14.4 kb), medium (M) segment (2.3 - 6.3 kb), and S segment (2 - 0.8 kb). The total genome length is 17,100 - 22,800 nucleotides, approximately 1 - 2% of the virion weight. The S fragment encodes the nucleocapsid protein, the M fragment encodes a large polypeptide (G), which is processed into G1 and G2 proteins, and the L fragment encodes the viral transcriptase (polymerase). Genetic variation exists in the S fragment structure among CCHFV strains from different regions. Phylogenetic analyses based on S RNA sequence indicate seven clades associated with geographic locations (24). Studies of CCHFV in ticks from Oman, the United Arab Emirates, Saudi Arabia, and Madagascar revealed two genetic classes of CCHFV, both observed in Iran. Accurate and convenient detection of CCHFV is crucial in developing countries. Therefore, a RT LAMP assay can serve as a valuable alternative to conventional methods.