1. Background
Pneumocystis jirovecii is an opportunistic fungal pathogen that primarily affects immunocompromised individuals, leading to Pneumocystis pneumonia (PCP), a significant cause of morbidity and mortality in this population. Originally classified as a protozoan, P. jirovecii has been recognized as a major contributor to respiratory infections in patients with conditions such as HIV/AIDS, hematological malignancies, and those undergoing immunosuppressive therapy (1, 2). The emergence of PCP as a critical health issue in the intensive care unit (ICU) setting underscores the need for timely and accurate diagnostics, particularly as the incidence of PCP continues to rise globally (3, 4).
The prevalence of P. jirovecii infections in critically ill patients has been reported to be alarmingly high, particularly among those with underlying conditions such as solid organ transplants, prolonged corticosteroid use, and hematological disorders (5). The ability to rapidly identify and genotype P. jirovecii in bronchoalveolar lavage (BAL) samples can provide essential information for clinicians, aiding in the selection of the most effective therapeutic regimens and improving patient management (6).
Traditionally, the diagnosis of PCP has relied on clinical symptoms, imaging studies, and histological examination of lung tissue or BAL fluid (7). However, these methods can be limited by their specificity and sensitivity, often leading to delayed treatment and poor patient outcomes (8). In contrast, molecular techniques, particularly polymerase chain reaction (PCR), have revolutionized the identification of P. jirovecii, allowing for rapid and precise detection of the organism's DNA in clinical samples (3, 9). This advancement is crucial in the ICU, where prompt diagnosis is essential for initiating appropriate therapy and improving survival rates (10).
The use of BAL samples for the molecular identification of P. jirovecii has gained prominence due to the high yield of pathogens retrieved from the lower respiratory tract (11). The BAL fluid not only provides a direct sample from the site of infection but also enhances the sensitivity of PCR assays compared to other sampling methods (12). Moreover, genotyping of P. jirovecii isolates can reveal genetic diversity, which may have implications for disease severity, treatment response, and the epidemiology of infections (13, 14). Understanding the genetic makeup of circulating strains is vital for monitoring potential outbreaks and resistance patterns (15).
Recent studies have demonstrated a correlation between specific genotypes of P. jirovecii and clinical outcomes in immunocompromised patients (16). For instance, certain genotypes have been linked to more severe manifestations of PCP, highlighting the importance of genotyping in predicting disease progression and tailoring therapeutic strategies (17). Furthermore, the emergence of novel genotypes in specific populations raises concerns regarding the adequacy of current treatment protocols and the necessity for ongoing surveillance (18).
2. Objectives
In this study, we aim to molecularly identify and genotype P. jirovecii in BAL samples of patients admitted to the ICU of eastern Iran, contributing to the growing body of literature that seeks to elucidate the complexities of this pathogen in critically ill populations. By employing advanced molecular techniques, we hope to enhance our understanding of the epidemiological trends and clinical implications of P. jirovecii infections in the ICU setting. The findings from this research could have significant implications for public health and clinical practice, particularly in optimizing treatment protocols and improving patient outcomes for those affected by PCP.
3. Methods
3.1. Patients
This descriptive study was carried out between May 2023 and January 2024, involving the collection of 100 BAL samples from patients admitted to the ICU at Waliasr Hospital, which is affiliated with Birjand University of Medical Sciences in Birjand, eastern Iran. The samples were obtained through random sampling methods. The sample size was determined based on findings from similar studies and the limited availability of samples.
A checklist was completed for each patient, containing demographic and clinical information, including age, sex, cause of hospitalization, and any underlying diseases. The study included adults aged 18 years and older who were admitted to the ICU. Participants were required to exhibit respiratory symptoms or have a suspected diagnosis of pneumonia. Additionally, BAL samples needed to be collected within 48 hours of their ICU admission. Patients with either a confirmed or suspected immunocompromised status were also eligible for inclusion.
Patients younger than 18 years were excluded from the study. The BAL samples that were contaminated or deemed unsuitable for molecular analysis, such as those with insufficient volume or improper storage, were also excluded. Furthermore, patients with known infections from other pathogens that could skew the study results, such as bacterial or viral pneumonia, were not included. Lastly, individuals admitted to the ICU for non-respiratory conditions and who did not exhibit respiratory symptoms were excluded from participation.
The Ethics Committee of Birjand University of Medical Sciences, Iran, approved the project (IR.BUMS.REC.1402.131).
3.2. Sampling
Samples were collected as mini-BAL samples from the endotracheal tube and placed in sterile Falcon tubes containing physiological saline. They were then sent to the medical microbiology laboratory at Birjand University of Medical Sciences for molecular testing.
3.3. DNA Extraction
Following the manufacturer's instructions, DNA was extracted using the Favorgen Biotech DNA Tissue Kit (Taiwan). Before nested PCR, the isolated DNA was stored in a -20°C freezer.
3.4. 18S rRNA Nested-Polymerase Chain Reaction
Nested PCR was performed to amplify the partial 18S rRNA gene of P. jirovecii, based on previous studies. In the first reaction, outer forward (5'- TTCGGGGCTTACTTTGGTC -3') and reverse (5'-GTAGTTAGTCTTCAATAAATCT-3') primers were used at an annealing temperature of 56°C. For the second reaction, inner forward (5'-AGGCCTACCATGGTTTCG-3') and reverse (5'-CTTCGGAGGACCGGGCCGT-3') primers were employed at an annealing temperature of 58°C, resulting in a product size of 330 bp (16, 19).
The cycling conditions for the first PCR reaction included an initial cycle of 5 minutes at 96°C, followed by 40 cycles of 96°C for 30 seconds, 60°C for 35 seconds, and 72°C for 40 seconds. The second PCR reaction included an initial cycle of 5 minutes at 96°C, followed by 30 cycles of 96°C for 30 seconds, 60°C for 35 seconds, and 72°C for 40 seconds (16).
The PCR products were analyzed by electrophoresis on 1.5% agarose gels containing 0.5 μg/mL DNA safe stain, and the bands were visualized under UV light using a transilluminator.
3.5. DNA Sequencing
The Applied Biosystems Big Dye terminator cycle sequencing kit and the 3130xl-ABI Genetic Analyzer were used to sequence the secondary PCR products. Chromas software was utilized to manually build and edit each sequence. The Basic Local Alignment Search Tool (BLAST) was employed to compare the results. Additionally, the trimmed nucleotide sequences were realigned using MEGA software version 5.00 with 1,000 replicates.
3.6. Statistical Analysis
The gathered data were analyzed using SPSS software (version 21, SPSS Inc., Chicago, IL) with the chi-square (χ2) test and logistic regression. Statistical significance was considered for P-values below 0.05.
4. Results
In this study, 100 patients admitted to the ICU of Waliasr Hospital in Birjand, eastern Iran, were examined using bronchoscopy. The group consisted of 48 males (48.0%) and 52 females (52.0%), aged between 20 and 85 years. The largest age group was 61 to 80 years, comprising 46.0% of the participants. The most common reasons for hospitalization were sepsis (47.0%) and lung disease (26.0%). The highest frequency of underlying conditions was observed in patients with hypertension (43.0%) and lung disease (39.0%). Regarding immunosuppression data, 8% of patients had various malignancies and 1% had organ transplants (Table 1).
| Demographic and Clinical Variables | All Patients | Patients with Pneumocystis jirovecii | |||
|---|---|---|---|---|---|
| No. (%) | No. (%) from Positive | P-Value | % from All | P-Value | |
| Gender | 0.356 | 0.360 | |||
| Female | 52 (52.0) | 6 (66.7) | 11.5 | ||
| Male | 48 (48.0) | 3 (33.3) | 6.3 | ||
| Age (y) | 0.392 | 0.4 | |||
| < 30 | 8 (8.0) | 0 (0) | 0 | ||
| 31 - 60 | 19 (19.0) | 1 (11.1) | 5.3 | ||
| 61 - 80 | 46 (46.0) | 6 (66.7) | 13.0 | ||
| > 81 | 27 (27.0) | 2 (22.2) | 7.4 | ||
| Cause of hospitalization | 0.302 | 0.315 | |||
| Sepsis | 47 (47.0) | 4 (44.4) | 8.5 | ||
| Lung disease a | 26 (26.0) | 0 (0) | 0 | ||
| Malignancy | 3 (3.0) | 0 (0) | 0 | ||
| Digestive diseases b | 6 (6.0) | 0 (0) | 0 | ||
| Poisoning | 3 (3.0) | 0 (0) | 0 | ||
| Brain diseases c | 9 (9.0) | 5 (55.6) | 55.5 | ||
| Heart diseases d | 2 (2.0) | 0 (0) | 0 | ||
| Other | 4 (4.0) | 0 (0) | 0 | ||
| Underlying diseases | 0.126 | 0.225 | |||
| Hypertension | 43 (43.0) | 6 (66.7) | 13.9 | ||
| Lung disease | 39 (39.0) | 0 (0) | 0 | ||
| Neurological diseases | 1 (1.0) | 0 (0) | 0 | ||
| Heart failure | 1 (1.0) | 0 (0) | 0 | ||
| Renal failure | 0 (0) | 0 (0) | 0 | ||
| Diabetes | 16 (16.0) | 3 (33.3) | 18.7 | ||
| Malignancy | 0 (0) | 0 (0) | 0 | ||
| Liver disease | 0 (0) | 0 0 | 0 | ||
| Dyslipidemia | 0 (0) | 0 0 | 0 | ||
| Hypothyroidism | 0 (0) | 0 0 | 0 | ||
| Organ transplant | 0 (0) | 0 0 | 0 | ||
| Total | 100 (100) | 9.0 | |||
a Lung disease: Chronic obstructive pulmonary disease (COPD), asthma, pneumonia, and interstitial lung disease (ILD).
b Digestive diseases: Gastritis, peptic ulcer disease, irritable bowel syndrome (IBS), and diarrheal diseases.
c Brain diseases: Stroke, Alzheimer, epilepsy, and migraine.
d Heart diseases: Coronary artery disease (CAD), heart attack, and arrhythmias.
The average duration of hospitalization, ICU stay, and intubation for the patients was 26.56 days, 24.36 days, and 23.63 days, respectively. The overall mortality rate for patients hospitalized in the ICU was 73%, and for patients with P. jirovecii, it was 4.1%.
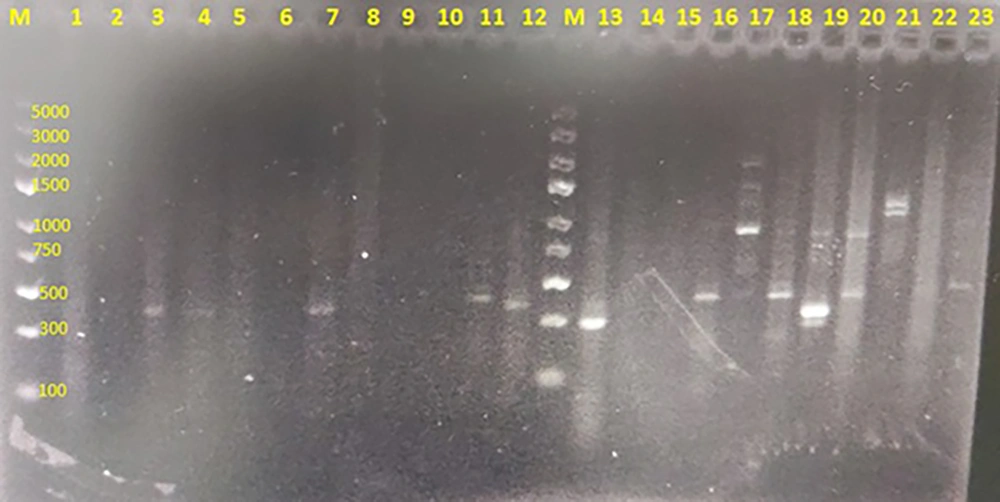
The nested-PCR technique targeting the 18S rRNA gene of P. jirovecii confirmed that 9 out of 100 BAL samples collected from patients admitted to the ICU at Birjand Waliasr Hospital in eastern Iran were positive (Figure 1). The majority of P. jirovecii positive cases were among individuals aged 61 to 80 years (66.7%), with no positive cases observed in patients under 30 years of age. Most of the positive cases were female (66.7%), while 33.3% were male. The primary reasons for hospitalization among the P. jirovecii positive cases were sepsis and neurological conditions, such as decreased consciousness, intracranial hemorrhage/intraventricular hemorrhage (ICH/IVH), and cerebrovascular accidents (CVA). Lung diseases were also a cause of hospitalization, though less frequently. The most common underlying conditions among the P. jirovecii positive cases were hypertension (66.7%) and diabetes (33.3%).
Agarose gel electrophoresis 1.5% of nested-polymerase chain reaction (PCR) products of Pneumocystis jirovecii partial 18SrRNA gene. Lanes 3, 4, 7, 11, 12, 16, 18, 20, and 23: Positive clinical samples having specific 330 bp bands representative of P. jirovecii. Lanes 1, 2, 5, 6, 8, 9, 10, 14, 15 and 22: No representative band indicating negative clinical samples. Lanes 13, 17, 19 and 21 have unspecific bands with unexpected size seen in some samples. Lanes M: A hundred bp DNA size marker.
Statistical analysis of the obtained data using the χ2 test showed that there was no significant relationship between positive cases of P. jirovecii and age, gender, reason for hospitalization, and underlying diseases (P > 0.05) (Table 1). Of the 9 patients with P. jirovecii, 7 received trimethoprim-sulfamethoxazole, with 4 recoveries and 3 deaths. The logistic regression results indicated that none of the predictors significantly influenced the outcome.
The analysis of the 18S rRNA gene sequence in five sequenced samples indicated that they belong to genotype III, which was identified as the dominant genotype. The sequences obtained from the 18S rRNA locus were compared with typical reference sequences from GenBank using a phylogenetic tree constructed with the maximum likelihood (ML) method, as shown in Figure 2. The accession numbers for the representative sequences in GenBank are PQ164802 to PQ164806.
5. Discussion
The findings from this study provide significant insights into the demographic and clinical characteristics of patients admitted to the ICU at Waliasr Hospital in Birjand, eastern Iran. With a total of 100 patients examined via bronchoscopy, our results highlight the prevalence of severe respiratory conditions and the corresponding underlying health issues that contribute to ICU admissions.
The study population consisted of 48 males (48.0%) and 52 females (52.0%), indicating a relatively balanced gender distribution. However, the largest age group was between 61 and 80 years, accounting for 46.0% of participants. This finding aligns with previous studies that have shown an increased incidence of severe respiratory illnesses in older adults, primarily due to age-related decline in immune function and the presence of comorbidities (20, 21).
The most common reasons for hospitalization were sepsis (47.0%) and lung disease (26.0%). Sepsis, a life-threatening organ dysfunction caused by a dysregulated host response to infection, remains a leading cause of ICU admissions globally (22). The high prevalence of lung disease among our patients is consistent with other research indicating that respiratory conditions are a significant contributor to morbidity and mortality in critically ill patients (23). Notably, the highest frequency of underlying conditions was observed in patients with hypertension (43.0%) and lung disease (39.0%). Hypertension is a well-documented risk factor for adverse outcomes in critically ill patients, as it is often associated with cardiovascular complications that can exacerbate respiratory failure (24). Similarly, lung diseases such as chronic obstructive pulmonary disease (COPD) and interstitial lung disease are known to increase the risk of respiratory infections and subsequent ICU admissions (25).
The average duration of hospitalization, ICU stay, and intubation were 26.56 days, 24.36 days, and 23.63 days, respectively. These prolonged durations reflect the severity of the patients' conditions and the complexity of their management in the ICU setting. Previous studies have reported similar lengths of stay, emphasizing the need for effective management strategies to optimize resource utilization in critical care environments (26).
The nested-PCR technique targeting the 18S rRNA gene of P. jirovecii confirmed that 9 out of 100 BAL samples were positive for this pathogen. The predominance of positive cases among individuals aged 61 to 80 years (66.7%) highlights the vulnerability of older populations to opportunistic infections, particularly in the context of immunocompromised states (27). The absence of positive cases in patients under 30 years of age further underscores this trend. Interestingly, most positive cases were female (66.7%), which might reflect gender differences in susceptibility to infection or the prevalence of underlying conditions such as autoimmune diseases that are more common in women (28).
The primary reasons for hospitalization among the P. jirovecii positive cases included sepsis and neurological conditions, with lung diseases also contributing, albeit less frequently. This finding is consistent with literature that identifies P. jirovecii pneumonia as a common complication in patients with sepsis and those with neurological impairments (29).
The analysis of the 18S rRNA gene sequence in five sequenced samples revealed that they belong to genotype III, identified as the dominant genotype. This genotype has been associated with increased virulence and resistance to treatment, emphasizing the importance of ongoing surveillance and research into the genetic diversity of P. jirovecii (30). Pneumocystis jirovecii genotype III is associated with higher virulence, particularly in immunocompromised individuals. This genotype may lead to more severe manifestations of PCP, increasing the risk of complications and mortality. Studies indicate that genotype III can elicit a stronger inflammatory response, which may contribute to lung damage and impaired gas exchange. The genetic factors underlying this increased virulence are still being investigated, but they may involve variations in surface proteins that enhance the organism's ability to evade the immune system. Additionally, patients infected with genotype III may experience a more rapid progression of symptoms, necessitating urgent medical intervention (31).
Monitoring genotype III prevalence in ICU patients provides valuable data on the epidemiology of P. jirovecii in the study area. Understanding the specific genotype can inform clinicians about potential variations in disease severity and treatment response. Studying genotype III contributes to the broader understanding of the pathogen's genetic diversity and its implications for virulence. Continuous analysis of BAL samples helps in establishing effective surveillance programs for early detection of outbreaks. Certain genotypes may exhibit resistance to standard treatments, such as trimethoprim-sulfamethoxazole, complicating management. Genotype III could influence the effectiveness of antifungal therapies, necessitating alternative treatment strategies. Variations in genotype may correlate with differences in disease severity and patient outcomes, impacting clinical decision-making. The host's immune response to specific genotypes can affect the clinical course of PCP, influencing recovery rates (32). Pneumocystis jirovecii genotype III has garnered attention due to its potential association with treatment resistance. Studies suggest that this genotype may exhibit variations in susceptibility to commonly used therapies, particularly trimethoprim-sulfamethoxazole, which is the standard treatment for PCP. Resistance can lead to treatment failures, resulting in prolonged illness and increased mortality rates in affected patients. The mechanisms behind this resistance are not fully understood, but genetic mutations may play a significant role. Additionally, the presence of genotype III in immunocompromised patients raises concerns about the effectiveness of prophylactic measures. Therefore, timely identification of this genotype in clinical settings is crucial for optimizing treatment strategies (33).
The phylogenetic comparison with reference sequences from GenBank further supports the need for a better understanding of the evolutionary dynamics of this pathogen. The phylogenetic tree of P. jirovecii specimens, alongside comparisons with other isolates, suggests that the isolates in this study share a common ancestor with those from Tehran and southwestern Iran. The close geographical relationship may have facilitated the movement and exchange of populations between these regions, contributing to this shared ancestry. Certain strains might exhibit similar ecological sequences within Iran, which could promote the selection and persistence of specific populations with common ancestors in the area. Notably, all isolates examined in this study were categorized as a distinct root, separate from P. carinii, which served as an outgroup.
Limitations of this study included the single-center design, available sample size, immunosuppression status, and unmeasured confounders.
5.1. Conclusions
In conclusion, this study highlights critical demographic and clinical characteristics of ICU patients in eastern Iran, emphasizing the significant burden of respiratory diseases and associated comorbidities. The detection of P. jirovecii in a subset of patients underscores the need for heightened awareness and appropriate diagnostic measures in this vulnerable population. Routine PCR screening and genotype surveillance are recommended for ICU patients. Future research should focus on exploring the implications of these findings for clinical practice and public health strategies aimed at improving outcomes for critically ill patients.