1. Context
Nonalcoholic fatty liver disease (NAFLD) encompasses two conditions characterized by excessive fatty deposition in the liver in the absence of excessive alcohol intake, namely hepatic steatosis and nonalcoholic steatohepatitis (NASH). Hepatic steatosis in the absence of alcohol intake occurs often in people with metabolic syndrome or any of its components, and insulin resistance. NASH involves lipotoxicity from accumulation of injurious lipid molecules, such as free fatty acids or free cholesterol, which are in turn associated with hepatic oxidative stress and recruitment of various cytokines leading to hepatic inflammation and fibrosis (1). Whereas the risk of cirrhosis development in hepatic steatosis over a 10-to 20-year period is lower than 4% (2), 5-8% of patients with NASH would develop liver cirrhosis within five years (3). Of note, NASH is currently recognized as a leading cause of cryptogenic cirrhosis (4). NAFLD is currently believed to affect approximately one-quarter of some populations (as high as 45% in some) and contributes to a striking proportion of liver disease burden in both Western and Asian countries (5-7). Therefore, NAFLD is a significant health hazard and economic burden. Differences in NAFLD prevalence, clinical profile and histological severity between different ethnic groups suggest a genetic contribution (5). This has prompted investigations into polymorphisms of several genes, including those involved in lipid handling, insulin signaling, oxidative stress and hepatic fibrosis. Among these, investigating gene polymorphisms of apolipoprotein C3 (APOC3) has recently attracted much interest (8). Two single nucleotide polymorphisms (SNPs) in the promoter region of the APOC3 gene (rs2854117 [–482C > T] and rs2854116 [–455T > C]) (which are in strong linkage disequilibrium with each other (8)) have been reported to be associated with hypertriglyceridemia, metabolic syndrome and coronary artery disease (9). More recently, these variants have been reported to be associated with the occurrence of NAFLD (8). Given the key role of APOC3 in NAFLD, several studies investigated the association between specific gene polymorphisms of APOC3 and NAFLD. Since many conflicting reports have been published to date on this issue, we performed a meta-analysis of all the relevant studies published in the literature to evaluate the association between gene polymorphisms of APOC3 and NAFLD.
2. Evidence Acquisition
2.1. Search Strategy
We aimed to identify published articles of all genetic association studies evaluating gene polymorphisms of APOC3 and NAFLD in humans, in all languages, up to June 2014. An electronic search was completed using PubMed, EMBASE, the Cochrane Library, and China National Knowledge Infrastructure. Different combinations of the keywords “APOC3” or “apolipoprotein C3” and “NAFLD” or “nonalcoholic fatty liver disease” were searched and not restricted by period. We also performed a full manual search from the bibliographies of selected papers. In addition, we contacted the authors of studies containing relevant information if there was lacking data necessary for the analysis. Unpublished data wasalso accepted if an abstract was available and further information was obtained from the authors.
2.2. Inclusion and Exclusion Criteria
In this meta-analysis, following inclusion criteria were set and reviewed by two independent investigators:
an independent case-control study;
studies with similar purpose and statistical methods;
studies providing enough information to calculate an odds ratio (OR);
Two SNPs of the APOC3 gene (rs2854117 [–482C > T] and rs2854116 [–455T > C]) were molecularly typed (high or low resolution level); and
fatty liver disease evaluated either by hydrogen magnetic resonance spectroscopy (H-MRS) (10) (NAFLD was defined as liver fat content of at least 55.6 mg triglycerides/g of liver tissue), liver ultra sonographic (US) examination indicative of fatty infiltration or liver biopsy.
Exclusion criteria were:
incomplete raw data;
repetitive reports (if more than one version of the same study was retrieved, only the most recent was used); and
materials and methods were not well-described or reliable. Assessment of study quality is of great importance for systematic reviews and meta-analyses; despite the fact, scoring methods are sometimes problematic (11) and may not accurately assess the quality measures of interest (12).
Therefore, we used reliability of patient selection, molecular typing method, and statistical analysis methods as quality variables.
2.3. Data Extraction
Two investigators (Zhang HY and Xin YN) independently evaluated study eligibility, graded quality, and extracted outcome data. Disagreements were resolved by consensus. For each study, information concerning the following characteristics of the subjects were collected: demographic information (age, sex, and ethnicity) and authors, publication year, journal, publication type and language, allele genotype, number of cases and controls, definitions used for NAFLD, NAFLD sample description, control sample description (if there was more than one control group, we chose the healthy group as the control group to minimize confounding factors).
2.4. Statistical Analysis
Homogeneity was calculated by Cochran’s Q test (α = 0.05). If the results of the Q test had no significant heterogeneity, the Mantel-Haenszel fixed-effect model (Peto method) was used for the combined data. If the results of the Q test had significant heterogeneity, the DerSimonian-Laird random-effects model (DL method) was used for the combination of data. A pooled OR was presented as a standard plot with 95% CIs. In the absence of heterogeneity, the two methods provided identical results. As a measure of association between NAFLD and APOC3 alleles, we combined ORs with 95% CIs stratified by gene subtypes of patients and controls in a study. Funnel plots were used to evaluate publication bias (13). We performed a sensitivity analysis to assess the stability of the results by sequential omission of individual studies. All P values presented were two-tailed. The analyses were performed using Revman 5.2 provided by the Cochrane Collaboration Internet.
| First Author, Year | References | Study/Center Description | Population Ethnicity Country | Study Design | N | Features and Patients Characteristics | Age of the Subjects | Liver Biopsy | Female, No. (%) |
|---|---|---|---|---|---|---|---|---|---|
| Petersen, 2010 | (8) | Prospective, multicenter | Mixed (Asian Indian/non-Asian Indian) | Population-based | 95 | Hepatic steatosis measured by H-MRS | Adults | NA | 0 (0) |
| Valenti, 2011 | (14) | Prospective, multicenter | Mixed (Italian/Newcastle, the UK) | Hospital-based | 1074 | Hepatic steatosis measured by biopsy | Adults and Pediatrics | Y | 301 (40) |
| Sentinelli, 2011 | (15) | Prospective, one center | Italian obese subjects Caucasians | Hospital-based | 170 | Hepatic steatosis measured by US | Adults | NA | NA |
| Hyysalo, 2012 | (16) | Prospective, one center | Finland | Population-based | 417 | Hepatic steatosis measured by H-MRS | Adults | NA | 220 (53) |
| Peter, 2012 | (17) | Prospective, multicenter | Caucasians Germany | Hospital-based | 330 | Hepatic steatosis measured by H-MRS | Adults | NA | 200 (61) |
| Verrijken, 2013 | (18) | Prospective, one center | Antverp (Belgium) Caucasians | Hospital-based | 287 | Hepatic steatosis measured by US and biopsy | Adults | Y | NA |
| Niu, 2014 | (19) | Prospective, one center | Chinese Han | Hospital-based | 799 | Hepatic steatosis measured by US | Adults | NA | 416 (52) |
a Abbreviations: H-MRS, hydrogen magnetic resonance (H-MR) spectroscopy; US, liver ultrasonographic examination; NA, not available.
3. Results
3.1. Search Results
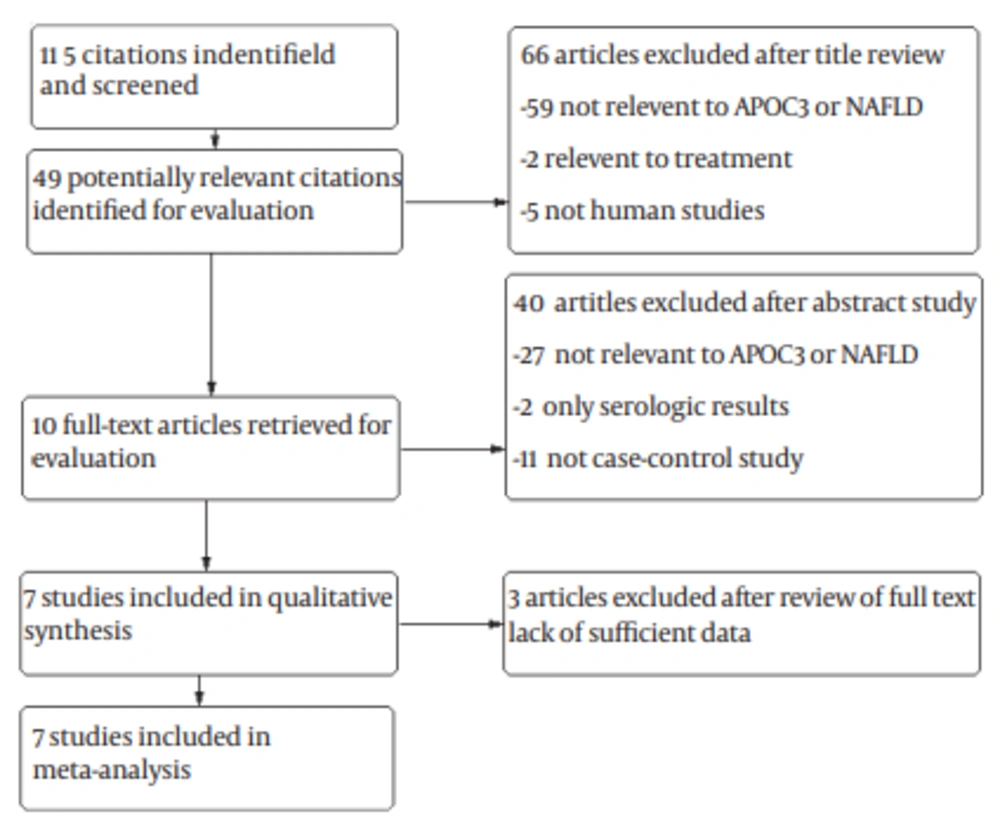
A total of 115 studies were retrieved based on the described search strategies. Ten eligible studies were identified for evaluation. Ultimately, three studies were excluded sue to insufficient data. Therefore, our final dataset for the meta-analysis (Figure 1) included seven studies (8, 14-19). The main features of the studies included in the meta-analysis are shown in Table 1. A total of 3172 subjects were included (1745 patients and 1427 controls). Two of these studies were conducted in Italy, (14, 15) and one each in the United States, (8) Finland, (16) Germany, (17) Belgium (18) and China (19). Five studies were hospital-based case-control studies, (14, 15, 17, 18-19) and the other population-based case-control studies (8, 16). Information about liver biopsy was available in two studies (14, 18). All the studies scored well for adequate descriptions of selection criteria, and availability of clinical data.
3.2. Association Between Gene Polymorphisms of APOC3 and NAFLD
Two SNPs of the APOC3 gene (rs2854117 [–482C > T] and rs2854116 [–455T > C]) were extracted from the studies to investigate their association with NAFLD.
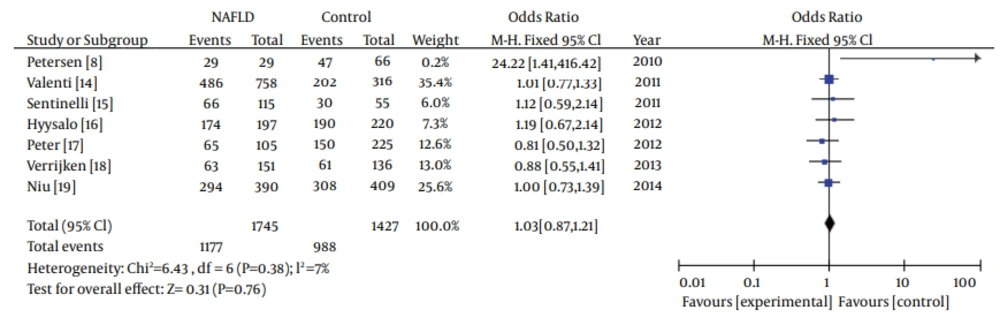
In the meta-analysis, the overall frequency of gene polymorphisms of APOC3 was 67.5% (1177/1745) in NAFLD, and 69.24% (988/1427) in controls. The heterogeneity test indicated that the variation of trial-specific ORs was not statistically significant (χ2 = 6.43, P = 0.38 and P > 0.1). The fixed-effect method was used to combine the results. The combined OR was 1.03 (95% CI: 0.87-1.21), which was not statistically significant (P = 0.76 and P > 0.05). In sensitivity analysis, exclusion of individual studies did not change this non-significant result. Statistics calculated for the study assessing the association between gene polymorphisms of APOC3 and NAFLD are shown in the forest plot (Figure 2).
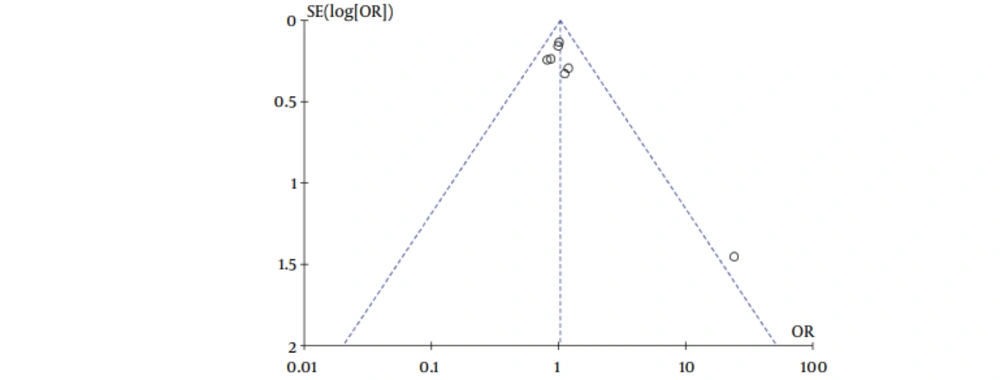
Our meta-analysis revealed no association between gene polymorphisms of APOC3 and NAFLD. Gene polymorphisms of APOC3 were not a risk factor for NAFLD. These analyses were based on the data from the study irrespective of population ethnicity. The funnel plot to detect publication bias of the study for APOC3 tends towards an asymmetrical shape (Figure 3), suggesting that publication bias might have affected the findings of our meta-analysis.
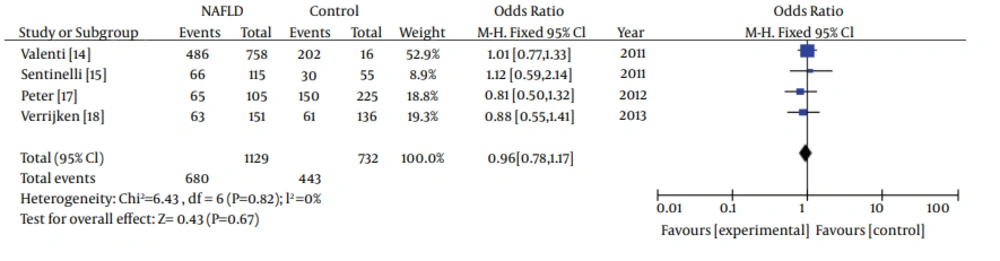
In subgroup analyses about ethnicity, heterogeneity test indicated that the variation of trial-specific ORs was not statistically significant (χ2 = 0.94, P = 0.82 and P > 0.1).The fixed-effect method was used to combine the results. The combined OR was 0.96 (95% CI: 0.78-1.17), which was not statistically significant (P = 0.67 and P > 0.05). Statistics calculated for the study assessing the association between gene polymorphisms of APOC3 and NAFLD in Caucasians are shown in the forest plot (Figure 4). Our subgroup analyses about ethnicity revealed no association between gene polymorphisms of APOC3 and NAFLD in Caucasians.
4. Discussion
The APOC3 protein is a component of triglyceride-rich lipoproteins, which inhibits lipoprotein lipase, which in turn hydrolyses triglycerides to generate free fatty acids before their uptake by muscle and adipose tissue, resulting in increased plasma triglyceride concentrations (20-22). Common mutations in the APOC3 promoter (-455T > C and -482C > T) have been associated with higher plasma triglycerides. These single nucleotide polymorphisms (SNPs) are located within an insulin responsive element in the APOC3 promoter and result in overexpression of APOC3 due to attenuated suppression by insulin. Numerous studies have reported associations of gene polymorphisms of APOC3 and higher plasma triglycerides (23). Whereas the physiological role and biological function of APOC3 in the liver are still unclear, we analyzed published evidence investigating the association between gene polymorphisms of APOC3 and NAFLD. Our primary purpose was to assess the effect of gene polymorphisms of APOC3 as a risk factor for developing NAFLD in different populations. To our knowledge, this was the first published meta-analysis to comprehensively investigate the association between gene polymorphisms of APOC3 and NAFLD. Studies concerning this possible association have been undertaken since 2010 (8). Asian-Indian men carrying at least one of the minor alleles of the rs3854116 (-455T > C) or the rs2854117 (-482C > T) SNPs in the APOC3 had higher liver fat content than homozygous carriers of both major alleles (8). In the same study, similar findings were observed for non-Asian Indian men. These data suggested that APOC3 may also be involved in the pathogenesis of fatty liver (8). In contrast, in a large study including three populations, SNPs were not associated with liver fat content (24). These contradictory findings necessitate further investigations on gene–environmental and gene-gene interactions of this genotype to determine hepatic steatosis (25). The summary OR for the association of gene polymorphisms of APOC3 and the risk of NAFLD was estimated as 1.03 with a 95% CI from 0.87 to 1.21. In our study, only English or Chinese publications were included in the analysis. A ‘Meta-analytical’ research on 29 meta-analyses investigating language bias provided evidence that OR values estimated in meta-analyses from non-English publications were on average 0.8-fold (95% CI, 0.7-1.0) of OR estimates from English-written publications (26). Therefore, even if we had not searched for non-English publications, this might have introduced only a small bias in the overall findings. Therefore, our language methodology is unlikely to have altered our main conclusions. However, the shape of the funnel plot seemed to be asymmetrical, suggesting that publication bias might have affected our findings. Several other points should be considered when interpreting the results of our study. First, three methods were used to detect liver fat content. Two studies (14, 18) performed a liver biopsy and the others used H-MRS (8, 16, 17) or ultrasound (15, 19). This may have introduced some heterogeneity in the diagnosis of NAFLD. Histology with standard staining, despite the possibility of false negative results, may be more precise than H-MRS and ultrasound and remains to be the reference method until present. However, most published data were based on H-MRS or ultrasound rather than liver biopsy. Secondly, because the information used in our research was based on data from observational studies, characteristics of each study population and different methodologies of these studies should be taken into account when interpreting the results of our analysis. For example, different inclusion criteria for selection of participants might have influenced the results of this research. In our study, only adults were included, whereas Valenti et al. (14) studied adults and children. Five studies (8, 14, 16, 17, 19) considered the proportion of women, while the other two studies (15, 18) did not consider gender ratio. Differences in age distribution, gender ratio and ethnicity could also be potential causes of variation in the study estimates. We also analyzed the ethnicity. Four studies (14, 15, 17, 18) reported subgroup analyses for Caucasians, but other ethnicities in selected researches were too little to perform subgroup analysis. Subgroup analyses about ethnicity suggested that gene polymorphisms of APOC3 were not a risk factor of NAFLD in Caucasians. Additionally, we tried to maximize our efforts to identify all relevant published studies in peer-reviewed journals, but it is possible to miss some. In conclusion, our analysis showed no association between gene polymorphisms of APOC3 and the risk of NAFLD. There may have been indications of possible publication bias and some heterogeneity in the methods used for assessing hepatic steatosis, with less pronounced associations in prospective studies than retrospective ones. Homogeneity of the methods for evaluating the degree of steatosis, common gender, age and ethnicity would be critical to confirm the absence of association and therefore lack of causal role of gene polymorphisms of APOC3 in patients with NAFLD. Given the importance of this issue, further prospective rigorous studies are warranted. Although, genetic predisposition to NAFLD may not be detectable until considering all other factors.