1. Background
Hepatocellular carcinoma (HCC) is one of the most prevalent malignant tumours worldwide, characterised by high incidence and mortality rates (1). Chronic infection with the hepatitis B virus (HBV) is recognised as a major risk factor for HCC. In recent years, increasing attention has been directed towards the hepatitis B virus X (HBx) protein and its pivotal role in the development, progression, and prognosis of HCC (2).
The HBx protein is a multifunctional protein encoded by the HBV genome and exerts a wide range of biological activities. It plays a critical role in viral replication, gene transcription, and the modulation of key intracellular signalling pathways (3). Studies indicate that HBx regulates various tumour-related cellular behaviours, including proliferation, chemoresistance, invasion, and metastasis (4). Specifically, HBx activates several signalling cascades and cytokines, such as p53, Wnt, and NF-κB pathways, thereby promoting tumour cell proliferation and metastasis (5, 6). Furthermore, HBx is closely associated with HCC recurrence and metastasis. Nevertheless, the precise molecular mechanisms by which HBx contributes to HCC pathogenesis remain incompletely understood and warrant further investigation (7).
Hepatic progenitor cells (HPCs) play a crucial role in liver development and regeneration. Keratin 19 (K19) is a specific molecular marker of HPCs. Although usually absent in normal liver tissue, K19 is expressed in approximately 20% of HCC cases. Cancer stem cell (CSC) characteristics are exhibited by K19-positive HCC cells, including multipotent differentiation capacity and distinct biological behaviours such as enhanced invasiveness, metastatic potential, and chemoresistance (8). Previous studies suggest that K19-positive HCC cells originate from HPCs or may arise from the dedifferentiation of malignant hepatocytes under continuous mutagenic stimulation. Additionally, the demethylation of CpG islands within the K19 promoter region may contribute to HCC development and progression. However, the precise mechanisms underlying the role of K19 in HCC, particularly its relationship with HBx, remain unclear (9).
Given the critical roles of both HBx and K19 in HCC, this study investigated their potential interaction and involvement in tumour invasion, metastasis, and recurrence. By modulating HBx and K19 expression in HCC cell lines, we examined their effects on key biological functions of HCC cells. We also analysed HBx and K19 expression patterns in HCC tissues and their correlations with clinicopathological characteristics and patient prognosis. These findings may provide novel insights and potential therapeutic targets for the diagnosis and treatment of HCC.
1.1. General Information
Patients diagnosed with HCC at our hospital between July 2020 and July 2025 were enrolled in this study. The inclusion criteria were as follows: (1) A documented history of HBV infection, with preoperative HBsAg or HBV-DNA positivity, and a postoperative pathological diagnosis confirming primary HCC; (2) age between 20 and 80 years; (3) absence of distant metastasis; (4) eligibility for curative surgical resection; and (5) no serious comorbid cardiopulmonary or systemic conditions. Exclusion criteria included: (1) Pathological diagnosis of metastatic liver cancer, cholangiocarcinoma, or other benign hepatic tumours; (2) palliative surgery with gross or microscopic residual tumour tissue; (3) severe cardiac, pulmonary, or neurological disease with an expected survival of less than six months; and/or (4) prior transarterial chemoembolisation before hepatectomy.
A total of 80 patients met the inclusion criteria and underwent curative surgical intervention. The cohort comprised 55 men and 25 women, aged 36 - 70 years, with a mean age of 53 years. Of these, 50 cases were classified as moderately to well-differentiated HCC, and 30 as poorly differentiated HCC. Hepatitis B infection was present in 53 cases, and 27 were negative for HBV markers. Serum alpha-fetoprotein (AFP) levels were > 400 (ρ/μg.L-¹) in 54 patients and < 400 (ρ/μg.L-¹) in 26 patients. Vascular invasion was observed in 51 patients and absent in 29. None of the patients had received surgical intervention, preoperative radiotherapy, or chemotherapy. All procedures and protocols were approved by the hospital’s ethics committee, and written informed consent was obtained from all participants.
2. Methods
Primary antibodies against HBx and cytokeratin 19 (CK19) were purchased from Servicebio (Wuhan, China). Human HCC cell lines SMMC-7721 and HepG2 were obtained from the cell bank of Zhongda Hospital Southeast University, Nanjing, Jiangsu Province, China. DMEM/F-12 medium was purchased from HyClone (USA), and fetal bovine serum (FBS) was obtained from Gibco (USA). Both the HBx and CK19 transfection reagents were purchased from TMO (USA). Primers were synthesised by GenePharma (Shanghai, China). Kits for RNA extraction were obtained from TIANGEN (Beijing, China), and complementary DNA synthesis kits and protein extraction kits were purchased from Bio-Rad (USA).
2.1. Immunohistochemical Staining
Immunohistochemical (IHC) staining was performed using the standard streptavidin-peroxidase method. Paraffin-embedded tissue sections were dewaxed and rehydrated through graded ethanol. Endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 5 - 10 minutes at room temperature. Antigen retrieval was conducted using microwave heating. Sections were then incubated with normal goat serum for 15 minutes at room temperature to block nonspecific binding, followed by overnight incubation with primary antibodies against p53 and survivin at 4°C. After washing with phosphate-buffered saline (PBS), biotinylated secondary antibodies were applied for 20 minutes at room temperature, then incubated with horseradish peroxidase (HRP)-conjugated streptavidin for another 20 minutes. Colour development was achieved using diaminobenzidine, and sections were counterstained with haematoxylin, rinsed with tap water, and mounted. Known HBx- and K19-positive HCC tissue sections were used as positive controls (10).
2.2. Interpretation of Immunohistochemical Staining Results
Cells exhibiting cytoplasmic staining from light yellow to dark brown were considered positive. Staining intensity was scored based on the predominant staining characteristics observed in the majority of cells, compared with background staining: No staining = 0 points, light yellow = 1 point, yellow-brown = 2 points, and dark brown = 3 points. The percentage of positively stained cells was evaluated across five randomly selected high-power fields per section and scored as follows: 0 - 9% = 1 point, 10 - 25% = 2 points, 26 - 50% = 3 points, and 51 - 100% = 4 points. For each field, both staining intensity and percentage scores were recorded. The final IHC score was calculated by multiplying the staining intensity score by the percentage score. Scores of 0 - 4 were classified as negative and > 4 as positive (10).
2.3. Cell Culture
Human HCC cell lines SMMC-7721 and HepG2 were revived from the cryopreserved cell bank of the Laboratory of Xuyi County People's Hospital. Revived cells were transferred to T25 culture flasks and maintained in complete DMEM/F12 medium supplemented with 10% FBS. Cells were incubated at 37°C in a humidified atmosphere containing 5% CO₂. Cells in the logarithmic growth phase were harvested for subsequent experiments (10).
2.4. Cell Transfection
After 24 hours of cell culture, the 6-well plates were inspected. Transfection was initiated when cells were in the logarithmic growth phase and reached approximately 60 - 70% confluency per well. Transfection was performed according to the manufacturer’s instructions.
Lentiviral (LV) vectors were constructed to overexpress HBx and K19 genes (LV-HBx and LV-K19), and the constructs were sequence-verified. These vectors were separately transfected into SMMC-7721 and HepG2 cells. The virus-transfection reagent mixtures were prepared according to the reagent protocol, added to the culture medium, gently mixed, and incubated at 37°C with 5% CO₂. Forty-eight hours post-transfection, cells were subjected to puromycin selection for three months to establish stably transfected lines. During this period, the puromycin-containing medium was refreshed every three days, resulting in stable cell lines overexpressing HBx and K19: LV-HBx-SMMC-7721, LV-HBx-HepG2, LV-K19-SMMC-7721, and LV-K19-HepG2.
To knock down K19 expression, a specific short hairpin RNA (shRNA) sequence targeting K19 was designed and cloned into an LV vector to generate the sh-K19 construct. A non-targeting shRNA sequence was also cloned into an LV vector as a negative control (NC). The sh-K19 and NC vectors were transfected into SMMC-7721 and HepG2 cells following the same procedure as for gene overexpression. Transfected cells underwent puromycin selection for three months to establish stable knockdown cell lines: sh-K19-SMMC-7721 and sh-K19-HepG2.
Following selection, the stably transfected cells were digested with trypsin-ethylenediaminetetraacetic acid, transferred to 15 mL centrifuge tubes, and centrifuged at 1,000 rpm for 5 minutes. After discarding the supernatant, the cell pellets were transferred to 1.5 mL LEP tubes for RNA extraction using TIANGEN kits (Beijing, China) (8).
2.5. Quantitative Real-time Polymerase Chain Reaction for Hepatitis B Virus X Protein and Keratin 19 Expression
Total RNA concentration was determined using a UV spectrophotometer (Model 752, Jinghua Instruments, Shanghai, China). Complementary DNA was synthesised using a Bio-Rad reverse transcription kit as instructed by the manufacturer. Polymerase chain reaction primers were designed using Primer Premier 5.0 (PREMIER Biosoft International). The primer sequences were as follows: (1) HBx: Forward: 5′-TAGGCTGTGCTGCCAACTGT-3′; Reverse: 5′-GGTCGTTGACATTGCTGAGA-3′; (2) CK19: Forward: 5′-TGCTGGATGAGCTGACTCTG-3′; Reverse: 5′-TGTTCAGCTCCTCAATCCGA-3′; (3) Internal Control (GAPDH): Forward: 5′-TTCTCCGAACGTGTCACGTTT-3′; Reverse: 5′-ACGTGACACGTTCGGAGAATT-3′.
Polymerase chain reaction was performed using the ABI 7500 real-time PCR detection system (Applied Biosystems, USA). The thermal cycling conditions were as follows: Initial denaturation at 94°C for 30 seconds, followed by 40 cycles of 94°C for 5 seconds, annealing at 50 - 60°C for 15 seconds, and extension at 72°C for 10 seconds. Relative mRNA expression levels were calculated using the 2-ΔΔCt method (11).
2.6. Western Blot Analysis of Hepatitis B Virus X Protein and Keratin 19 Protein Expression
Total cellular protein was extracted using a Bio-Rad protein extraction kit following the manufacturer’s instructions. Protein concentrations were determined by bicinchoninic acid assay. Proteins were denatured by boiling and stored at -80°C. For Western Blot (WB) analysis, 60 μg of total protein from each sample was separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes.
Membranes were blocked with 5% non-fat milk in TRIS-buffered saline with 2% Tween 20 (TBST) for 2 hours at room temperature and incubated overnight at 4°C with the following primary antibodies: Anti-HBx (1:100), anti-K19 (1:100), and anti-GAPDH (1:2000). After washing three times with TBST, membranes were incubated with HRP-conjugated secondary antibodies (1:4000) for 1 hour at room temperature. Following additional TBST washes, protein bands were visualised using enhanced chemiluminescence, and images were developed using photographic film. Band intensities were quantified using a BandScan 5.0 image analysis system (Epson, Japan) (7).
2.7. Wound Healing Assay
The HCC cell lines SMMC-7721 and HepG2 were cultured in complete DMEM/F12 medium supplemented with 10% FBS and incubated at 37°C in a humidified atmosphere containing 5% CO₂ until the cells reached 80 - 90% confluence. Subsequently, a sterile 200 μL pipette tip was used to create a straight vertical scratch across the monolayer to simulate a wound. Care was taken to ensure consistent scratch width across all samples. Detached cells and debris were gently removed by washing the monolayer three times with PBS. Cells were then maintained under standard culture conditions (37°C, 5% CO₂), and wound closure was monitored and imaged at 0, 24, and 48 hours using a phase-contrast microscope.
The initial and residual wound widths were measured using image analysis software (ImageJ), and wound-healing rates were calculated using the following formula:
Wound healing rates were compared among different treatment groups to assess variations in cell migratory capacity (7).
2.8. Invasion Assay
Cell invasion ability was assessed using a Transwell chamber system (3422, Corning) with an upper cell compartment and a lower culture medium reservoir. To mimic the extracellular matrix (ECM), the upper chamber membrane was pre-coated with Matrigel (354277, Corning). To minimise the influence of serum on invasive behaviour, SMMC-7721 and HepG2 cells were serum-starved for 24 hours before the assay, then resuspended at a density of 1 × 10⁵ cells/mL in serum-free medium and seeded into the upper chamber. The lower chamber was filled with culture medium containing 10% FBS as a chemoattractant.
The chambers were incubated at 37°C in 5% CO₂ for 24 - 48 hours to allow cells to invade through the Matrigel-coated membrane. At the end of incubation, invaded cells on the lower surface were fixed with 4% paraformaldehyde and stained with crystal violet. Five random microscopic fields were selected per membrane, and the number of invaded cells was counted under a light microscope. The average number of invaded cells per group was calculated, and differences in invasion capacity among different treatment groups were statistically analysed (7).
2.9. Tumoursphere Formation Assay
The HCC cell lines SMMC-7721 and HepG2 were cultured in serum-free DMEM/F12 medium supplemented with B27, epidermal growth factor, and basic fibroblast growth factor to promote sphere formation. Cells were seeded at a low density (1 × 10³ cells/mL) into ultra-low attachment culture dishes to prevent cell aggregation. Cultures were maintained at 37°C in a 5% CO₂ incubator, with medium changes every 3 - 4 days.
Tumoursphere formation was observed and photographed under a fluorescence microscope on days 0, 3, and 7. The tumoursphere formation rate was calculated using the following formula:
Sphere formation capacities were compared across different treatment groups.
2.10. Patient Follow-up
Postoperative follow-up was conducted for all 80 patients with HCC, beginning on the day of hospital discharge after surgery and ending on 1 July 2025. During follow-up, patients were monitored for overall survival (OS) and recurrence or metastasis of HCC. Overall survival was defined as the time from the date of surgery to death from any cause or to the last follow-up date. Recurrence-free survival (RFS) was defined as the time from the date of surgery to the first diagnosis of HCC recurrence or metastasis. For patients without recurrence or metastasis by the end of follow-up, the final follow-up date was considered the RFS endpoint.
2.11. Statistical Analysis
All statistical analyses were performed using SPSS 17.0 (SPSS Inc.). The chi-square (χ²) test was used to evaluate associations between categorical variables, and Spearman’s rank correlation coefficient was calculated to assess correlation strength. Univariate survival analysis was conducted using the Kaplan-Meier method, and differences in OS and RFS were compared using the log-rank test. Survival curves were plotted accordingly. A significance level of α = 0.05 was applied. One-way analysis of variance was used to compare means among multiple groups. A two-sided P-value < 0.05 was considered statistically significant.
3. Results
3.1. Efficient Overexpression and Knockdown of Hepatitis B Virus X Protein and Keratin 19 in Hepatocellular Carcinoma Cell Lines
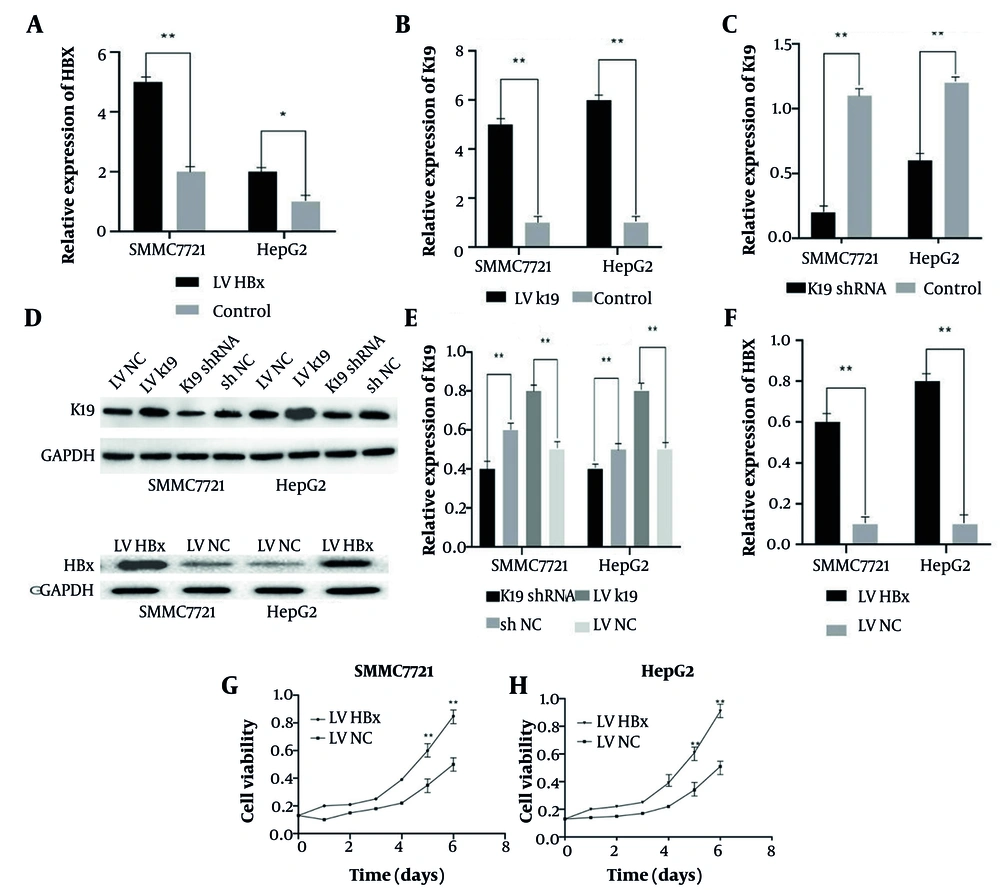
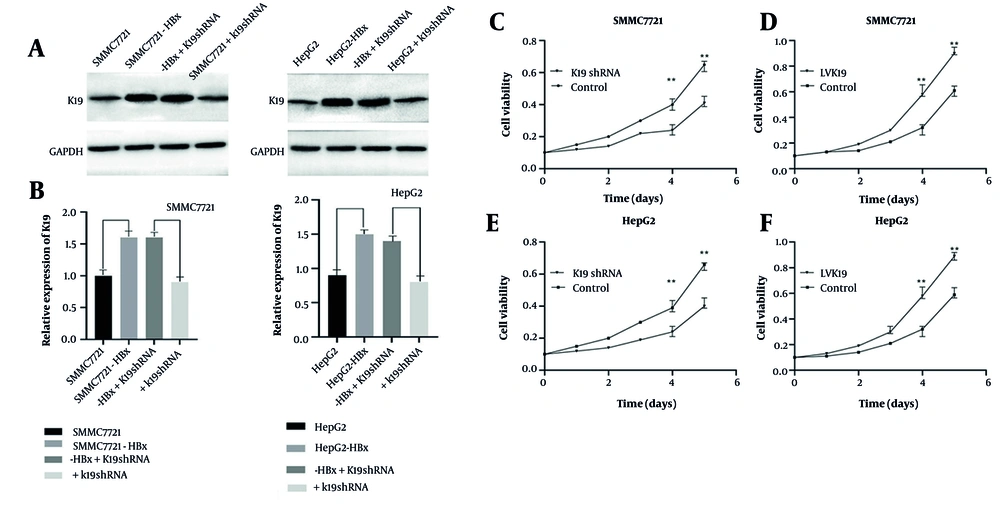
The HCC cell lines SMMC-7721 and HepG2 were successfully transfected to upregulate HBx and K19 mRNA expression and downregulate K19 protein expression. The quantitative real-time polymerase chain reaction (qRT-PCR) results showed significantly elevated HBx and K19 mRNA levels in the overexpression groups compared with controls (n = 3, P < 0.05; Figure 1A and B). In the knockdown groups, K19 mRNA expression was significantly decreased (n = 3, P < 0.05; Figure 1C). The WB analysis confirmed the qRT-PCR findings: The mRNA levels of HBx and K19 were significantly upregulated in the overexpression groups (n = 3, P < 0.05; Figure 1D - F), whereas K19 protein levels were significantly reduced in the knockdown groups (n = 3, P < 0.05; Figure 1D and E). In order to clarify the effect of HBx on the proliferation ability of HCC cells, SMMC-7721 and HepG2 cells were stably transfected with LV-HBx and LV-NC. In the MTT assay after cell transfection, the proliferation ability of HCC cells in the HBx-transfected group was significantly enhanced compared with the control group (P < 0.01; Figure 1G and H). The above results indicate that HBx promotes the proliferation ability of HCC.
Detection of transfection effects. qRT-PCR was used to detect hepatitis B virus X protein (HBx) gene expression level in SMMC-7721 and HepG2 cells after HBx transfection (n = 3, P < 0.01) (A). qRT-PCR was used to detect the keratin 19 (K19) gene list of SMMC-7721 and HepG2 cells transfected with Lentiviral (LV) (n = 3, P < 0.05) (B). qRT-PCR was used to detect the expression level of K19 gene in SMMC-7721 and HepG2 cells transfected with K19 shRNA (n = 3, P < 0.01) (C). Western-blot to detect the expression levels of K19 and HBx protein in SMMC-7721 and HepG2 cells after transfection with K19 shRNA, Lentiviral (LV) K19, and LV HBx (D). Western-blot statistical analysis of K19 protein expression levels in SMMC-7721 and HepG2 cells transfected with K19 shRNA and LV K19 (n = 3, P < 0.01) (E). Western-blot statistical analysis of the expression level of HBx protein in SMMC-7721 and HepG2 cells transfected with LV HBx (n = 3, P < 0.01) (F). Effect of HBx transfection on the proliferation ability of SMMC-7721 (n = 3, P < 0.01) (G). Effect of HBx transfection on HepG2 proliferation (n = 3, P < 0.01) (H). * P < 0.05, * * P < 0.01.
3.2. Hepatitis B Virus X Protein Overexpression Enhances Hepatocellular Carcinoma Cell Migration in Wound-Healing Assays
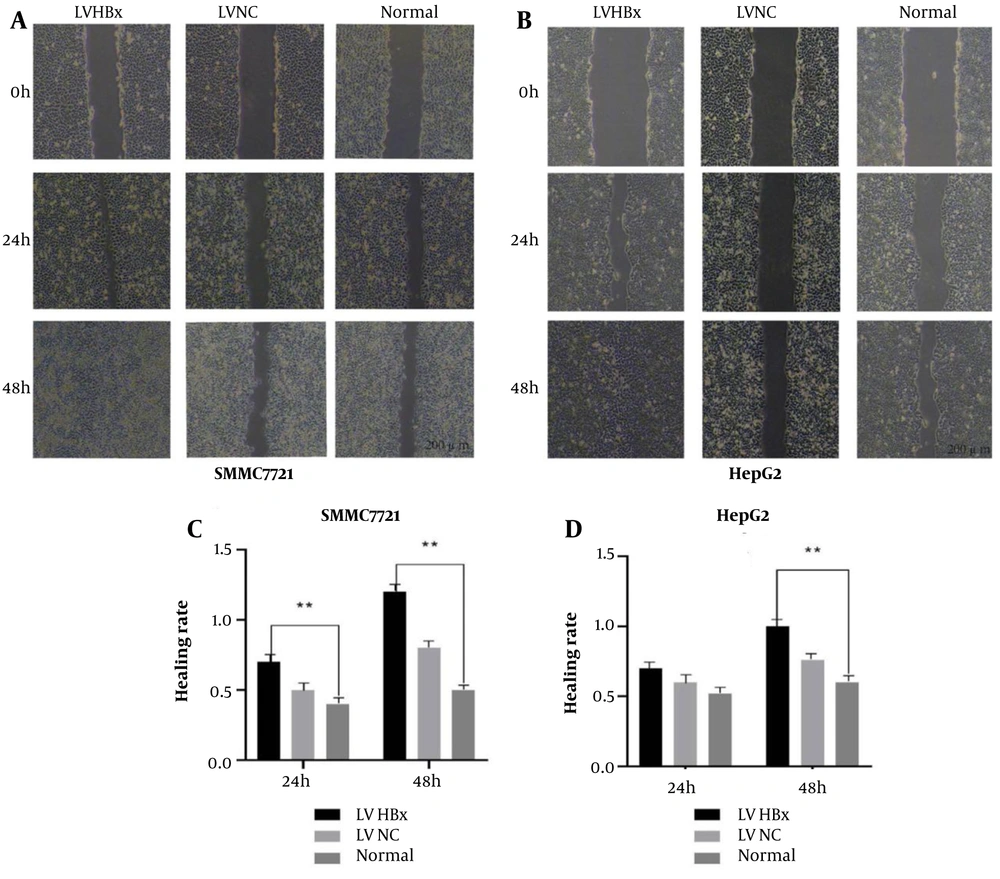
In wound-healing assays, both cell lines exhibited significantly higher wound-healing rates in the LV-HBx group compared with the control (LV-NC) and normal (untransfected) groups (all P < 0.05; Figure 2A and B). At 48 hours, the wound-healing rate was significantly higher in the HBx-overexpression groups than in the control group (P < 0.01; Figure 2C and D). Given the positive correlation between wound-healing rate and cell migratory capacity, these results demonstrate that HBx promotes the migration of HCC cells.
The effect of Lentiviral (LV) hepatitis B virus X protein (HBx) transfection on the migration ability of hepatocellular carcinoma (HCC) cells. Scratch test to detect the effect of LV HBx transfection on the migration ability of SMMC-7721 (A). Scratch test to detect the effect of LV HBx transfection on HepG2 migration ability (B). Scratch experiment to analyze the effect of LV HBx transfection on migration ability of SMMC-7721(n = 3, P < 0.01) (C). Scratch test to analyze the effect of LV HBx transfection on HepG2 migration ability (n = 3, P < 0.01) (D). (** P < 0.01)
3.3. Hepatitis B Virus X Protein Overexpression Increases Hepatocellular Carcinoma Invasion Capacity
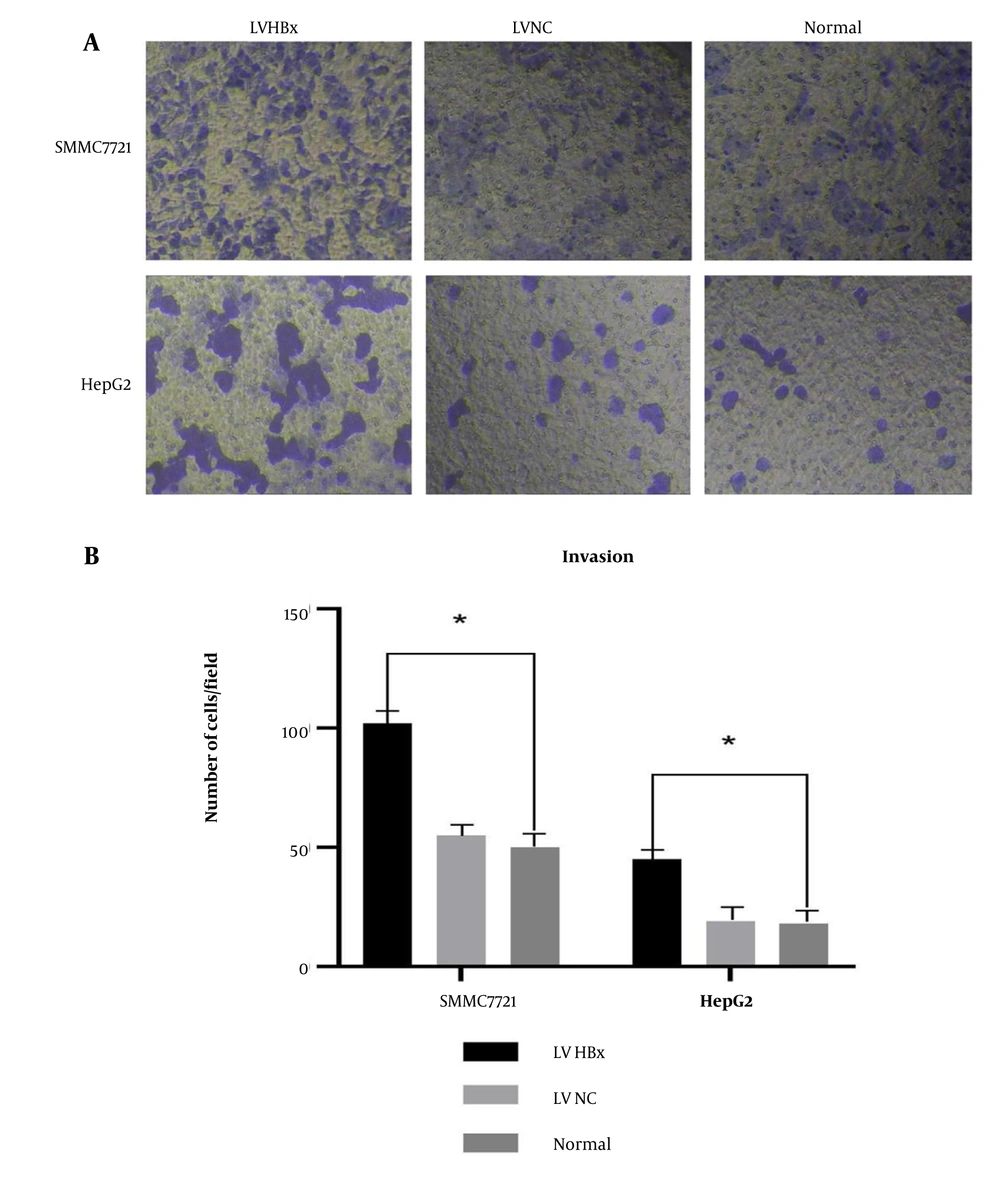
The quantitative analysis of invaded cells revealed that significantly more cells penetrated the Matrigel-coated in the HBx-overexpression group (LV-HBx) than in the control (LV-NC) and normal groups (P < 0.05; Figure 3A and B). This finding suggests that HBx enhances the invasive ability of HCC cells.
The effect of Lentiviral (LV) hepatitis B virus X protein (HBx) transfection on the invasion ability of hepatocellular carcinoma (HCC) cells. Invasion experiment to detect the effect of LV HBx transfection on the invasion ability of HCC cell lines. (Magnification: 40X) (A). Invasion experiment to analyze the effect of LV HBx transfection on the invasion ability of HCC cells (n = 3, P <0.05) (B). (* P < 0.05)
3.4. Hepatitis B Virus X Protein Promotes Tumoursphere Formation
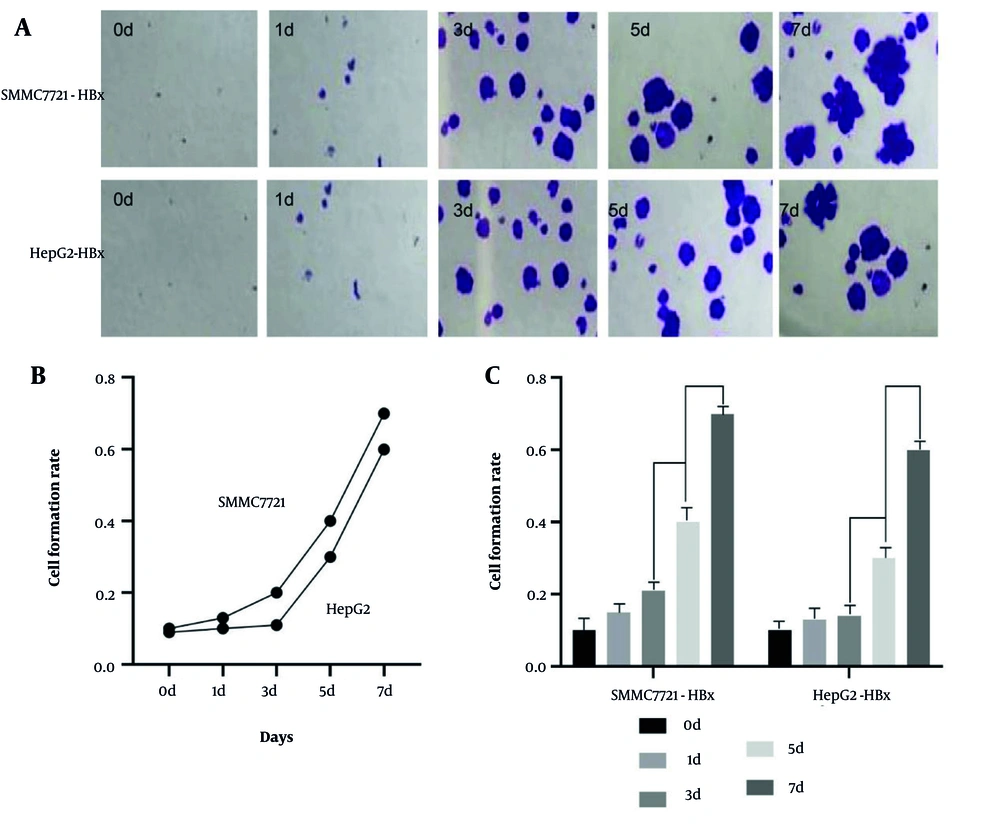
Fluorescence imaging from day 0 to day 7 showed a time-dependent increase in sphere formation in the HBx-overexpression group. The quantitative analysis demonstrated a significantly higher tumoursphere-formation rate in the HBx-overexpression group between days 3 and 7 compared with controls (P < 0.05; Figure 4A - C). These results support the role of HBx in promoting the tumoursphere-forming ability of HCC cells.
The effect of Lentiviral (LV) hepatitis B virus X protein (HBx) transfection on the ball forming ability of hepatocellular carcinoma (HCC) cells. Ball formation experiment to detect the effect of LV HBx transfection on the ball forming ability HCC cell lines. (Magnification: 40X) (A). Ball formation to analyze the effect of LV HBx transfection on the invasion ability of HCC cells (n = 3, P < 0.05) (B, C).
3.5. Hepatitis B Virus X Protein Up-regulates Keratin 19 Protein Expression
The WB analysis revealed that SMMC-7721 and HepG2 cells exhibited the highest levels of K19 protein expression among all groups (n = 3, both P < 0.05; Figure 5B). Interestingly, when K19 was knocked down using K19 shRNA in the parental cell lines, K19 expression was significantly reduced; however, in HBx-overexpressing cells treated with K19 shRNA, the reduction in K19 expression was only modest and not statistically significant, contrasting with the previous knockdown results (n = 3, P < 0.05; Figure 5A, B). These findings suggest that HBx may upregulate K19 expression and that HBx may promote HCC cell invasion and migration via K19 regulation.
Detect the changes of keratin 19 (K19) protein by regulating hepatitis B virus X protein (HBx) and K19. Observe the changes of related K19 protein in SMMC-77721 and HepG2 cells by regulating HBx and K19 in western-blot experiment (A). In the western-blot experiment, Lentiviral (LV) HBxSMMC-7721, LV HBxHepG2 cell analysis down-regulated K19 and caused changes in K19 protein (n = 3, P <0.05) (B). Effect of transfection with K19 shRNA on the proliferation ability of SMMC-7721 (n = 3, P < 0.01) (C, D). Effect of transfection of K19 shRNA on proliferation of HepG2 cells (n = 3, P < 0.01) (E, F).
In order to clarify the effect of K19 on the proliferation ability of HCC cells, SMMC-7721 and HepG2 cell lines were stably transfected with K19 shRNA and LVK19. In the MTT assay after cell transfection, the K19 down-regulation group showed a significant decrease in the proliferation ability of HCC cells compared with the control group (P < 0.01; Figure 5C and Figure 5D). Compared with the control group, the K19 up-regulation group showed a significant improvement in the proliferation ability of HCC cells (P < 0.01; Figure 5E and F). The above results showed that K19 promoted the proliferation ability of HCC.
3.6. Keratin 19 Mediates Hepatitis B Virus X Protein-Driven Migration and Invasion
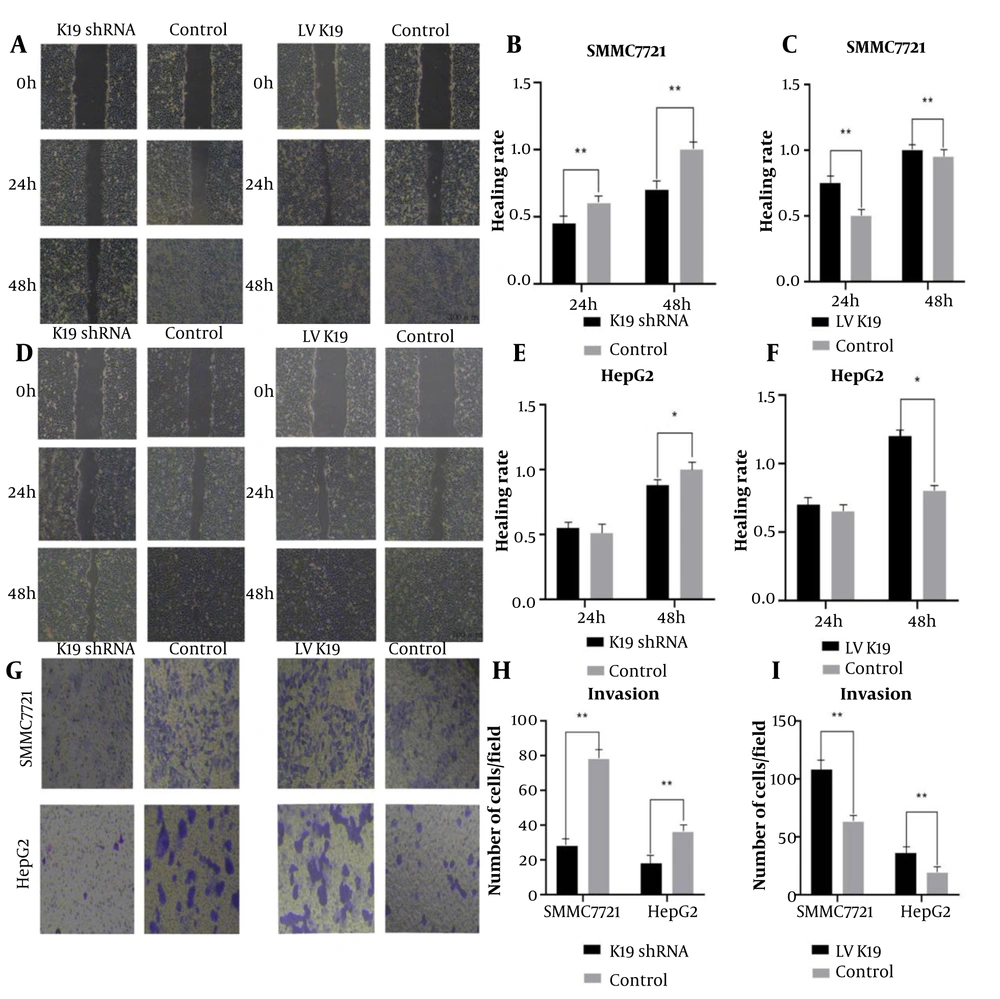
Stable HBx overexpression was confirmed in LV-HBx-SMMC-7721 and LV-HBx-HepG2 cells after 3 days of puromycin selection by qRT-PCR and WB analysis. To investigate whether K19 mediates HBx-induced cellular effects, these HCC cell lines were further transfected with K19 overexpression or knockdown constructs (i.e. LV-K19, K19 shRNA, NC). In wound healing assays, the K19 knockdown group showed significantly reduced wound-healing rates compared with the control group in both LV-HBx-SMMC-7721 and LV-HBx-HepG2 cells (P < 0.05; Figure 6A, D). In the SMMC-7721 cell line, K19 knockdown led to a significantly lower wound-healing rate at 48 hours compared with the control group (P < 0.01; Figure 6B). Conversely, K19 overexpression resulted in a significantly enhanced wound-healing rate compared with that in the control group (P < 0.05; Figure 6A, C, D, E, F).
Hepatitis B virus X protein (HBx) enhances the migration and invasion ability of hepatocellular carcinoma (HCC) cells through keratin 19 (K19). The scratch experiment was used to detect the effect of K19 shRNA and Lentiviral (LV) K19 transfection on the migration ability of stable transfected cells LV HBxSMMC-7721 (A). The healing rate of scratch area was used to analyze the migration ability of K19 shRNA transfected LV HBxSMMC-7721 stable transfected cells. (N = 3, P < 0.01) (B). The effect of transfection of LV K19 on the migration ability of stable transfected cells LV HBxSMMC-7721 (n = 3, P < 0.01) (C). Scratch test was used to detect the effect of K19 shRNA and LV K19 transfection on the migration ability of stable transfected cells LV HBxHepG2 (D). The effect of K19 shRNA transfection on the migration ability of stable transfected cells LV HBxHepG2 was analyzed by the scratch area healing rate (n = 3, P < 0.05) (E). The effect of LVK19 transfection on the migration ability of stable transfected cells LV HBxHepG2 was analyzed by the healing rate of scratch area (n = 3, P < 0.05) (F). Invasion test Effect of K19 shRNA and LV K19 transfection on the invasion ability of stable transfected cells LV HBxSMMC-7721, LV HBxHepG2 (Magnification: 40X, n = 3, P < 0.01) (G). Analysis of K19 shRNA transfection on stable transfected cells LV by cell counting HBxSMMC-77721, the effect of LV HBxHepG2 invasion ability (n = 3, P < 0.01) (H). Transfection of LV K19 to stable transfected cells LV HBxSMMC-7721 by cell count analysis LV HBxHepG2 invasive ability (n = 3, P < 0.01) (I). * P < 0.05, ** P < 0.01
In the Transwell invasion assay, K19 overexpression led to a significant increase in the number of invasive cells in the LV-HBx-SMMC-7721 and LV-HBx-HepG2 cell lines compared with the control groups (P < 0.01; Figure 6G and I). In contrast, K19 knockdown significantly reduced the number of invading cells compared with controls (P < 0.01; Figure 6G and H).
3.7. Positive Correlation Between Hepatitis B Virus X Protein and Keratin 19 Protein Levels in Human Hepatocellular Carcinoma Tissues
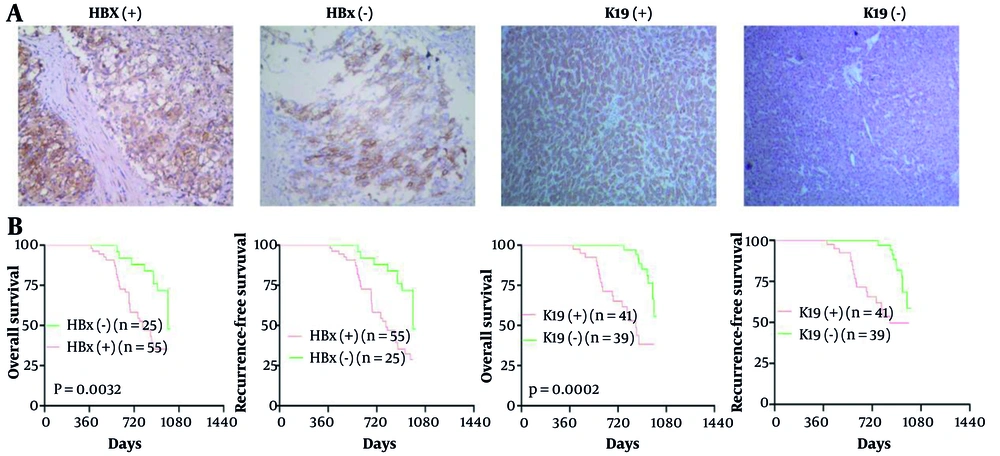
The positive expression of HBx and K19 is primarily observed as brown-yellow cytoplasmic staining in HCC cells. Expression of the HBx protein was positive in 68.75% (55/80) and negative in 31.25% (25/80) of the 80 HCC tissue samples, and K19 expression was positive in 51.25% (41/80) and negative in 48.75% (39/80). The differences were statistically significant (χ² = 5.10, P < 0.05). Among HBx-positive HCC tissues, the rate of K19 positivity was 65.45% (36/55), whereas in HBx-negative HCC tissues, K19 positivity was 34.55% (19/55), indicating a statistically significant difference (χ² = 7.04, P < 0.01). The rank correlation analysis further revealed a positive correlation between HBx and K19 expression in HCC tissues (r = 0.8333, P < 0.01) (Table 1 and Figure 7). Kaplan–Meier analysis showed worse 5-year overall survival and recurrence-free survival in patients with HBx-positive (OS 31% vs 48%; RFS 27% vs 48%; P = 0.0032) and K19-positive tumours (OS 29% vs 54%; RFS 46% vs 56%; P = 0.0002) (Figure 7B).
The expression levels of hepatitis B virus X protein (HBx) and keratin 19 (K19) protein in tumor specimens of hepatocellular carcinoma (HCC) patients and the survival curves of HBx and K19 expression on overall survival (OS) and recurrence-free survival (RFS) in HCC patients. The expression levels of HBx and K19 protein in tumor specimens from patients with HCC were evaluated by immunohistochemical (IHC) analysis (SP × 100) (A). Kaplan-Meier analysis was conducted to assess the survival curves of HBx and K19 expression on OS and RFS in patients with HCC (B).
| HBx Expression | K19 | χ2 | P-Value | |
|---|---|---|---|---|
| Positive | Negative | |||
| Positive | 36 | 19 | 7.04 | < 0.01 |
| Negative | 5 | 20 | ||
Abbreviation: HCC, hepatocellular carcinoma.
3.8. Association of Hepatitis B Virus X Protein/Keratin 19 Expression with Aggressive Clinicopathological Features
Statistical analysis showed no significant association between HBx or K19 protein expression and patient age, sex, or Child–Pugh liver function classification (all P > 0.05). Similarly, liver cirrhosis was not significantly correlated with K19 expression (P > 0.05) but was significantly associated with HBx expression (P < 0.05), potentially reflecting HBV infection. In contrast, HBx and K19 protein expression were both significantly associated with several clinical factors, including preoperative serum AFP levels, HBV infection status, histological differentiation of HCC, vascular invasion, and tumour–node–metastasis (TNM) stage (* P < 0.05, ** P < 0.01) (Table 2).
| Clinicopathological Characteristics | K19 Protein Expression | χ²-Value | P-Value | HBx Protein Expression | χ²-Value | P-Value | ||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |||||
| Gender | 1.84 | > 0.05 | 2.75 | > 0.05 | ||||
| Male (n = 55) | 31 | 24 | 41 | 14 | ||||
| Female (n = 25) | 10 | 15 | 14 | 11 | ||||
| Age (y) | 1.63 | > 0.05 | 0.93 | > 0.05 | ||||
| ≤ 50 (n = 26) | 16 | 10 | 16 | 10 | ||||
| > 50 (n = 54) | 25 | 29 | 39 | 15 | ||||
| Child-pugh liver function classification | 0.64 | > 0.05 | 0.22 | > 0.05 | ||||
| A | 26 | 28 | 31 | 16 | ||||
| B | 15 | 11 | 4 | 4 | ||||
| C | 0 | 0 | 20 | 5 | ||||
| Liver cirrhosis | 2.47 | > 0.05 | 11.65 | < 0.05 a | ||||
| Absent (n = 28) | 11 | 17 | 26 | 2 | ||||
| Present (n = 52) | 30 | 22 | 29 | 23 | ||||
| AFP (ng/mL) | 24.31 | < 0.01 b | 3.98 | < 0.05 a | ||||
| ≥ 400 (n = 54) | 38 | 16 | 41 | 13 | ||||
| < 400 (n = 26) | 3 | 23 | 14 | 12 | ||||
| HbsAg status | 21.65 | < 0.01 b | 63.02 | < 0.01 b | ||||
| Positive (n = 53) | 37 | 16 | 52 | 1 | ||||
| Negative (n = 27) | 4 | 23 | 3 | 24 | ||||
| Tumor differentiation level | 9.37 | <0.01 b | 4.75 | < 0.05 a | ||||
| Moderately/Well-differentiated (n = 50) | 19 | 31 | 30 | 20 | ||||
| Poorly differentiated (n = 30) | 22 | 8 | 25 | 5 | ||||
| Vascular invasion | 5.71 | < 0.05 a | 6.45 | < 0.05 a | ||||
| Present (n = 51) | 21 | 30 | 30 | 21 | ||||
| Absent (n = 29) | 20 | 9 | 25 | 4 | ||||
| TNM stage | 7.02 | < 0.01 b | 4.62 | < 0.05 a | ||||
| I-II (n = 52) | 21 | 31 | 40 | 12 | ||||
Abbreviations: HCC: hepatocellular carcinoma; AFP, alpha-fetoprotein; TNM, tumor node metastasis classification.
a P < 0.05.
b P < 0.01.
4. Discussion
Primary HCC is a common malignancy characterised by high aggressiveness (11). According to the most recent global cancer statistics published by the WHO (12), the annual incidence of new cancer cases worldwide has reached 18 million. Among these, liver cancer ranks sixth in incidence and fourth in cancer-related mortality, and it is the second leading cause of cancer-related death in men globally. Accumulating evidence suggests that HBx is closely associated with HCC recurrence and metastasis (13-18). In this study, we demonstrated that HBx significantly enhances the migration, invasion, and tumoursphere-forming capacity of HCC cells through upregulation of K19 expression. Both HBx and K19 were frequently overexpressed in HCC tissues, and their expression levels were positively correlated. Clinically, positive HBx and K19 expression was associated with aggressive tumour characteristics, including poor differentiation, vascular invasion, advanced TNM stage, and elevated AFP levels. Kaplan–Meier analysis revealed that patients with HBx- or K19-positive tumours had significantly worse OS and RFS compared with those with negative expression. These findings suggest that HBx promotes HCC progression and recurrence through K19-mediated enhancement of CSC-like properties, and that HBx and K19 may serve as valuable prognostic biomarkers and potential therapeutic targets in HBV-related HCC.
As a surface marker of liver CSCs (LCSCs), K19 is closely associated with aggressive tumour behaviour (19, 20). Stem cell-like properties are exhibited by K19-positive HCC cells, including multipotent differentiation potential and distinct biological behaviour. In HCC, K19 expression is primarily found in biliary progenitor cells and HPCs, where it plays a promotive role in cancer development and progression. Although HBx has previously been shown to regulate LCSCs and promote recurrence and metastasis, recent studies suggest that K19-positive HCC cells may originate from HPCs. Another hypothesis proposes that K19 expression in human HCC may result from the dedifferentiation of malignant hepatocytes due to continuous mutagenic stimulation. From an epigenetic perspective, CpG island demethylation in the K19 gene promoter region has been implicated in promoting HCC development and progression. Cells expressing K19 not only exhibit progenitor cell markers but also possess invasive and metastatic potential, as well as resistance to chemotherapy (21). Research further suggests that K19-positive HCC cells exhibit CSC-like features, including epithelial-mesenchymal transition (EMT) and TGF-β/Smad signalling cascade activation. Epithelial-mesenchymal transition is characterised by the loss of epithelial markers and the acquisition of mesenchymal traits, and is now recognised as a key mechanism underlying invasion and migration of cancer cells (including HCC cells) (22). Hepatocellular carcinoma cells largely retain the expression of cytokeratins consistent with their epithelial origin, whereas HPCs, which originate from biliary epithelial cells in the periportal or ductular regions of the liver, play a critical role in liver repair and regeneration and frequently express K19 in early stages (23).
Liu et al. (24) reported that the transfection of HBx into Huh7 cells significantly enhanced their invasive and migratory capabilities. Moreover, when SK-Hep1 cells infected with Ad-HBx were xenografted into immunodeficient mice, tumours in the Ad-HBx group exhibited substantial accumulation of infiltrating stromal cells and massive production of intratumoural ECM after one month, which contributed to the marked elongation of tumour cells and loss of tissue integrity. These findings indicate that HBx enhances the invasive and metastatic potential of HCC in the mouse model.
In this study, HBx expression was upregulated through LV transfection, and HCC cell lines with relatively high HBx expression levels were selected through qRT-PCR and WB analysis to investigate the functional impact of HBx on the invasive and metastatic behaviour of HCC cell lines. Previous studies have shown that HBx can activate stemness-associated genes such as octamer-binding transcription factor 4 (OCT4), both in vitro and in vivo, thereby promoting cell migration, drug resistance, and metastasis or recurrence in HBV-related HCC (25, 26). Consistent with these findings, our results demonstrated that HBx overexpression significantly enhanced both the invasive and migratory capacities of HCC cell lines. In the wound healing assay, clear differences between the control and HBx-overexpression groups were observed as early as 24 hours, with statistically significant acceleration of wound healing, which became even more pronounced by 48 hours. Similarly, in the Transwell invasion assay, HBx upregulation markedly increased the invasive capacity of HCC cells. These findings revealed the potential association between HBx overexpression and HBV-related HCC.
From our tumoursphere formation assay, we observed that HBx overexpression significantly enhanced the sphere-forming capacity of HCC cells. This finding is consistent with previous reports that HBx overexpression induces stem cell-like traits in HepG2 cells (27), characterised by increased invasion, migration, and sphere-forming capacities. The enhanced invasive and metastatic behaviour of HBx-overexpressing HCC cells further supports the hypothesis that HBx may contribute to HCC development and progression by modulating LCSCs. Thus, targeting HBx may suppress HBx-induced hepatocarcinogenesis and delay the progression of HBV-related HCC.
Recent studies have also implicated HBx in promoting CSC generation during HCC development (28). Both in vitro and in vivo, HBx has been shown to activate a range of stemness-associated proteins, including β-catenin, epithelial cell adhesion molecule, Klf-4, Nanog, and OCT4 (29). Clarifying the role of HBx may help deepen our understanding of postoperative recurrence, metastasis, and the emergence of chemoresistance in HCC, and may inform the development of more effective surgical, chemotherapeutic, and targeted modalities.
By jointly investigating HBx and K19, two key regulators of cancer stemness in HCC, we provide novel insights into HCC diagnosis, treatment, and recurrence prevention. Notably, K19-positive HCC exhibits CSC-like features and is consistently linked to poor prognosis (30). Thus, K19 expression may serve as an indicator of poor prognosis in patients with HCC.
Previous studies have reported that K19 is a surface marker of liver CSCs and plays a role in maintaining stem-like phenotypes, thereby enhancing the invasive and metastatic capabilities of tumour cells in vivo (31-33). In our study, we modulated K19 expression through upregulation and downregulation in LV-HBx-transfected SMMC-7721 and HepG2 cells to evaluate its effects on cellular migration and invasion. Overexpression of K19 significantly enhanced both invasive and migratory capacities in HBx-stable cell lines, whereas K19 knockdown suppressed these functional behaviours. These findings are consistent with previous clinical observations, suggesting that K19 is a critical regulator of malignant biological behaviour in HCC.
Our WB analysis further revealed that K19 protein expression was upregulated following HBx overexpression. Introduction of K19 shRNA significantly reduced K19 levels in parental HCC cell lines but failed to do so in HBx-overexpressing cells, indicating that K19 expression is likely regulated by HBx. Shen et al. (34) reported that HBx activates the Wnt/β-catenin signalling pathway, a fundamental axis in stem cell biology. Given that K19 is a stemness-associated marker, we hypothesise that HBx may promote HCC cell invasion and metastasis by upregulating K19 through activation of the Wnt/β-catenin signalling pathway.
Chen et al. (35) used IHC analysis to investigate the clinical significance of HBx protein expression in 110 cases of HCC. Their findings demonstrated that high HBx expression was significantly associated with elevated AFP levels and vascular invasion, suggesting that HBx may serve as an independent prognostic biomarker for poor outcomes in HCC. In our study, the positive and negative expression rates of HBx in HCC tissues were 68.75% and 31.25%, respectively. In patients with preoperative AFP levels > 400 ng/mL, concurrent HBV infection, poorly differentiated tumours, vascular invasion, and advanced TNM stage, HBx positivity was significantly higher (all P < 0.05). These results indicate the close relationship between HBx expression and aggressive tumour behaviour in HCC and identify HBx as a clinically meaningful biomarker for predicting poor prognosis in patients with HCC.
In patients with HCC, K19 positivity has similarly been reported as a useful predictor of recurrence following hepatic resection. Tsuchiya et al. (36) found that patients with K19-positive HCC experienced intrahepatic recurrence after radiofrequency ablation, with 6 out of 10 cases showing recurrence within 12 months. These findings suggest that K19 positivity is a major risk factor for recurrence and early relapse. In our study, the Kaplan-Meier survival analysis revealed that patients with HBx- or K19-positive HCC had significantly worse OS and RFS compared with those with negative expression of HBx or K19. Cox regression analysis further confirmed that HBx positivity was associated with an increased risk of poor prognosis, with hazard ratios for OS and RFS of 2.965 and 3.223, respectively. The expression of K19 was also identified as a significant prognostic factor for both OS and RFS in our cohort.
4.1. Conclusions
In summary, our findings suggest that HBx-mediated upregulation of K19 may promote invasion and metastasis in HBV-related HCC and is associated with unfavourable OS and RFS outcomes. Targeting HBx and K19 may represent a promising therapeutic strategy in the treatment of HCC, and assessment of their expression levels may serve as valuable prognostic biomarkers for the clinical management of HCC.