1. Background
Phenylalanine ammonia-lyase (PAL) is a key enzyme in the phenylpropanoid pathway, catalyzing the conversion of L-phenylalanine to cinnamic acid, thereby initiating the biosynthesis of a wide range of secondary metabolites, including flavonoids, anthocyanins, and procyanidins (1, 2). These compounds have attracted considerable attention due to their strong antioxidant activity, anti-inflammatory effects, and potential roles in the prevention of cardiovascular diseases (3).
Peach (Prunus persica), as one of the most important fruits in temperate regions, is rich in oligomeric procyanidins (OPCs). Beyond their nutritional properties, these compounds can interact with key cardiac proteins, potentially contributing to the prevention or modulation of cardiac disorders. A major focus in this context is the investigation of OPCs’ potential interaction with myosin binding protein C (MYBPC3), cardiac type, a protein whose mutations or functional alterations are directly associated with hypertrophic cardiomyopathy (HCM) (4).
In recent years, bioinformatic approaches and molecular modeling have emerged as powerful tools for identifying ligand-protein interactions and predicting the molecular pathways influenced by natural compounds (5-7). Molecular docking enables the identification of binding sites, interaction energies, and structural features of active sites, thereby facilitating the design of natural molecule-based therapeutics.
In this study, the physicochemical properties, as well as the secondary and tertiary structures of the PAL protein extracted from peach, were first analyzed. Subsequently, the interactions of three major procyanidins (B1, B2, and C1) with the human MYBPC3 protein were investigated using molecular docking. Additionally, cleft, pore, and tunnel analyses were conducted to determine potential pathways for ligand access to binding sites.
2. Objectives
The aim of this research is to identify the potential of peach-derived OPCs as natural molecules influencing MYBPC3 function and to evaluate their prospective applications in improving or preventing cardiac diseases such as HCM.
3. Methods
3.1. Extraction and Selection of Peach Phenylalanine Ammonia-Lyase Gene Sequence
The nucleotide sequence of the PAL gene from peach (P. persica) was retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov). The gene ID in NCBI is 18784865, and the corresponding mRNA sequence is XM_007220568.2. The translated protein sequence, with the ID XP_007220630.2, was used as a reference for structural and functional analyses (8).
3.2. Analysis of Physicochemical and Structural Properties of Phenylalanine Ammonia-Lyase Protein
The PAL protein sequence was submitted to the ExPASy ProtParam tool (https://web.expasy.org/protparam) to calculate key physicochemical parameters, including molecular weight, isoelectric point (pI), Instability Index, Aliphatic Index, and the grand average of hydropathicity (GRAVY) (9).
3.3. Tertiary Structure Prediction of Phenylalanine Ammonia-Lyase Protein
The three-dimensional structure of P. persica PAL1 protein was modeled using the Swiss-Model server (https://swissmodel.expasy.org). The final model was selected based on the Global Model Quality Estimation (GMQE) and QMEAN scores (10). Model quality was further evaluated using MolProbity (11), and the refined structure was prepared for structural and functional analyses using PyMOL software (12).
3.4. Identification and Optimization of Oligomeric Procyanidins
Three compounds — procyanidin B1, procyanidin B2, and procyanidin C1 — were selected based on previous studies. Their chemical structures were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). When necessary, 3D models were generated and optimized using the minimum energy algorithm based on the MMFF94 force field. The final structures were saved in PDB format (Table 1) (13).
| Compound | CID | Molecular Formula | Molecular Weight (g/mol) |
| Procyanidin B1 | 11250133 | C30H26O12 | 578.50 |
| Procyanidin B2 | 130556 | C30H26O12 | 578.50 |
| Procyanidin C1 | 169853 | C45H38O18 | 866.80 |
3.5. Analysis of Structure and Function of Human Myosin Binding Protein C Gene
The protein sequence of human MYBPC3 was retrieved from UniProt (accession code: Q14896), and its predicted three-dimensional structure was obtained from the AlphaFold Protein Structure Database (AF-Q14896-F1-v4). This model was subsequently prepared for molecular interaction analysis with OPC compounds (7, 14).
3.6. Molecular Docking of Oligomeric Procyanidins with Myosin Binding Protein C
The interactions between OPCs and MYBPC3 protein were investigated using AutoDock 4.2 software (15). Both the protein and ligand structures were converted to PDBQT format, and the target region, including the protein’s active sites, was defined. The results were analyzed in terms of binding energy (ΔG), interaction types, and locations, including hydrogen bonds and van der Waals interactions.
4. Results
4.1. Peach Phenylalanine Ammonia-Lyase Protein
4.1.1. Extraction and Selection of Phenylalanine Ammonia-Lyase Gene Sequence
The nucleotide sequence of the PAL1 gene from peach (P. persica) was retrieved from the NCBI database. The gene ID in NCBI is 18784865, and the corresponding mRNA sequence is XM_007220568.2. The translated protein sequence, with the ID XP_007220630.2, was selected as a reference for structural and functional analyses.
4.1.2. Physicochemical Properties of Phenylalanine Ammonia-Lyase Protein
The PAL protein sequence consists of 719 amino acids, with a molecular weight of 78,241.91 Da and a pI of 6.05, indicating a slightly acidic nature. The number of positively charged residues (Arg + Lys) is 72, while negatively charged residues (Asp + Glu) are 83, suggesting a balanced charge distribution. The protein Instability Index was calculated as 30.46, classifying PAL as stable. The Aliphatic Index of 88.07 and a GRAVY value of -0.227 indicate relative thermal stability and hydrophilicity. The predicted half-life of the protein is 30 hours in mammalian reticulocytes, over 20 hours in yeast, and more than 10 hours in E. coli, reflecting high stability across different systems.
4.1.3. Tertiary Structure Prediction of Phenylalanine Ammonia-Lyase Protein
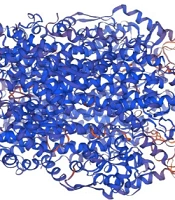
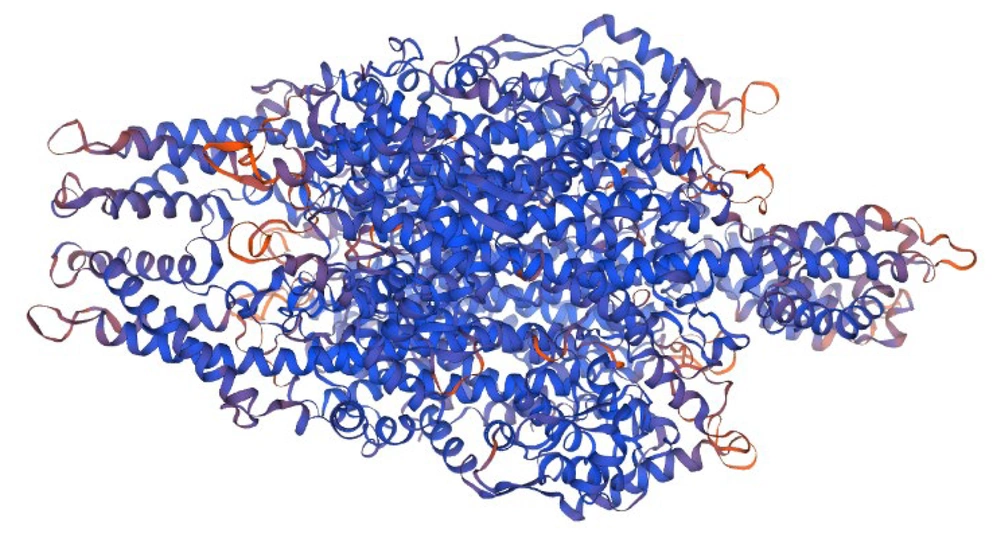
The three-dimensional structure of peach PAL1 protein was modeled using Swiss-Model. The final model was predicted as a homo-tetramer (Figure 1), and its quality was confirmed with GMQE = 0.86 and QMEANDisCo Global = 0.83 ± 0.05. MolProbity assessment indicated a MolProbity Score of 1.63, Clash Score of 0.92, and Ramachandran Favoured regions of 93.42%, demonstrating high model quality. This model is suitable for structural analyses and docking with OPC compounds.
4.2. Identification, Extraction, and Optimization of Oligomeric Procyanidin Structures
In this study, three representative compounds from the OPC family — procyanidin B1, procyanidin B2, and procyanidin C1 — were selected as representatives of OPCs. The selection was based on previous studies highlighting their presence in peach and their potential roles in biosynthetic pathways and biological activities. The three-dimensional structures of procyanidin B1 (CID: 11250133) and procyanidin B2 (CID: 130556) were directly retrieved from the PubChem database, whereas the structure of procyanidin C1 (CID: 169853) was initially obtained in 2D format and subsequently converted to a 3D model using Avogadro software. All compounds were geometrically optimized and energy-minimized using the MMFF94 force field. The optimized models retained active phenolic groups and B-type inter-unit linkages in all compounds, which are essential for biological activity and interactions with target proteins. The final structures were saved in PDB format and are ready for use in molecular docking studies.
4.3. Structural and Functional Analysis of Human Myosin Binding Protein C Gene
The MYBPC3 gene, a key regulator of cardiac function and sarcomere structural stability, was investigated in this study. The human protein sequence was retrieved from UniProt (accession code: Q14896), and its predicted three-dimensional structure was obtained from the AlphaFold Protein Structure Database (AF-Q14896-F1-v4). The model exhibited high pLDDT values, indicating a high confidence in the protein structure prediction. Physicochemical analysis revealed that MYBPC3 consists of 1,274 amino acids with an approximate molecular weight of 140 kDa. The protein contains multiple Ig-like C2-type and fibronectin type-III (FnIII) domains, which play critical roles in maintaining sarcomere structural stability. ProtParam calculations indicated a pI of approximately 6.05 and a long predicted half-life, reflecting relative stability under physiological conditions. Secondary structure prediction using SOPMA showed that the protein comprises a combination of α-helices, β-sheets, and random coils, with approximately 27.3% α-helix, 22.6% β-sheet, and 45.1% random coil. This structural diversity suggests protein flexibility and its capability to interact with other sarcomere components (Table 2).
| Secondary Structure Type | No. (%) of Residues |
|---|---|
| Alpha helix (H) | 27 (27.30) |
| Extended strand (E) | 23 (22.60) |
| Beta turn (T) | 5 (5.00); estimated |
| Random coil (C) | 45 (45.10) |
The three-dimensional model of the MYBPC3 protein highlighted the C-terminal domains (C5 - C10) in functional regions and was prepared for subsequent analyses, such as molecular docking. This model provides a foundation for investigating protein interactions with small ligands and assessing potential molecular effects on protein function.
4.4. Molecular Docking Results of Procyanidin Ligands with Myosin Binding Protein C
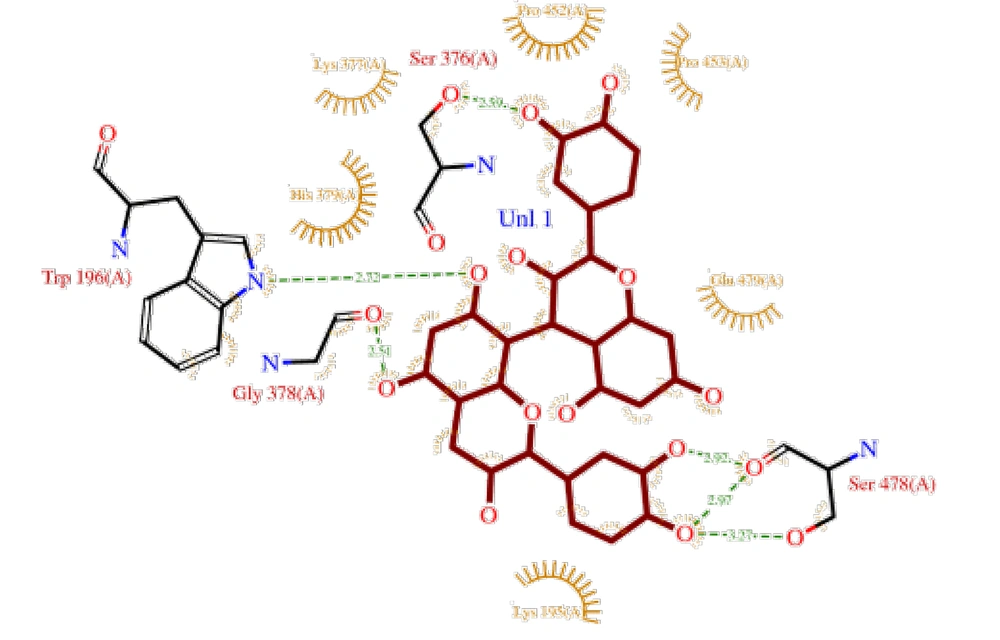
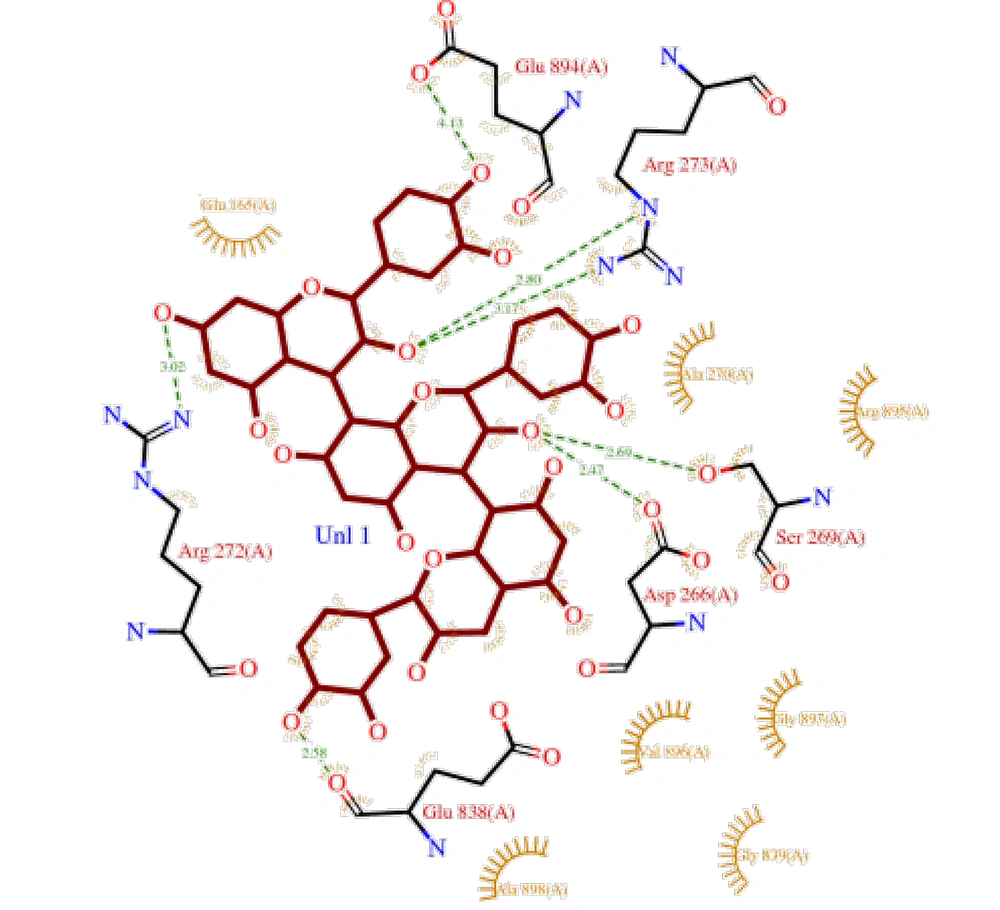
The interactions of three compounds — procyanidin B1, procyanidin B2, and procyanidin C1 — with MYBPC3 protein were investigated using molecular docking with AutoDock 4.2 software. The protein and ligand structures were converted to PDBQT format, and the target region included the active sites and potential binding regions of the protein. Procyanidin B1 exhibited a binding energy of -7.39 kcal/mol and an inhibition constant (Ki) of 3.78 µM, indicating a relatively strong ligand-protein interaction (Figure 2). Interaction analysis revealed that this ligand forms contacts with Glu48, Phe51, Asp32, and Tyr25 residues within cleft region 2. Cleft, pore, and tunnel analyses showed that MYBPC3 possesses multiple potential pathways for ligand accommodation, with procyanidin B1 occupying a semi-buried site within one of these regions.
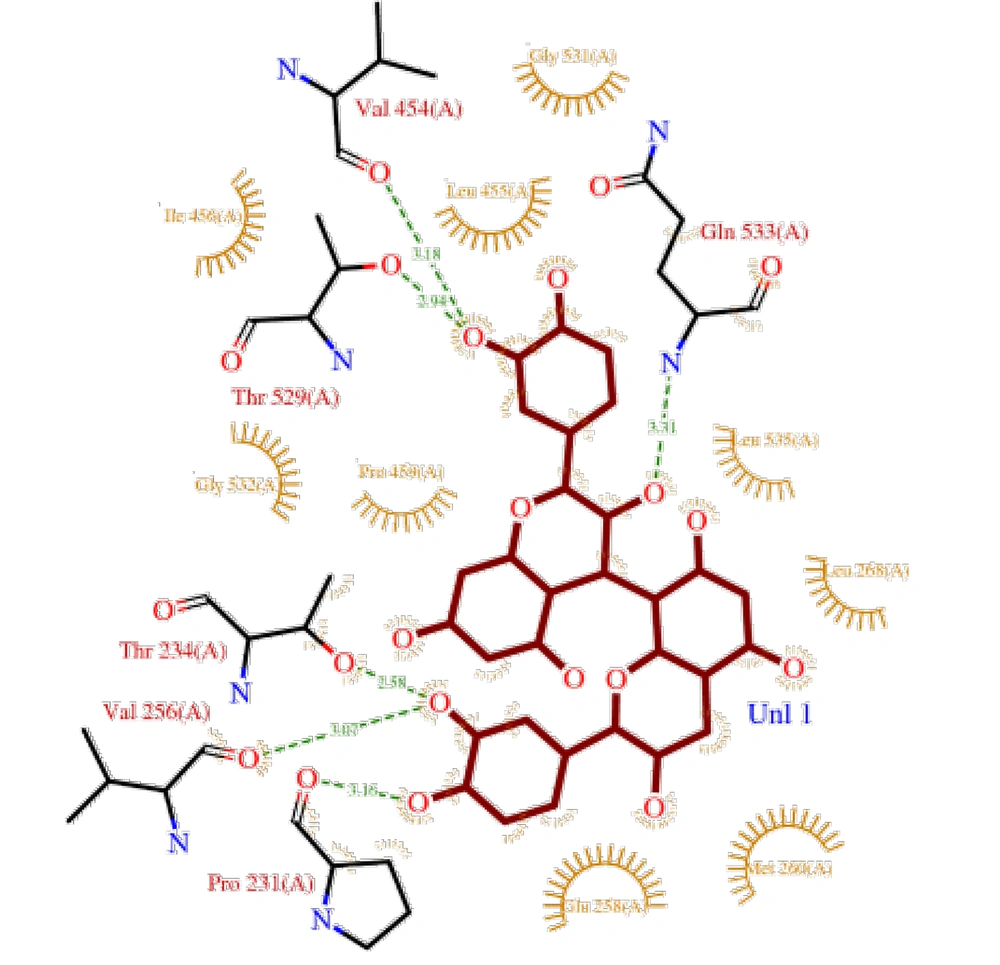
Procyanidin B2 also exhibited a significant interaction with the protein, with a binding energy of -7.12 kcal/mol and a Ki of 5.23 µM (Figure 3). This ligand interacted with aromatic and polar residues within cleft region 2. Similar to procyanidin B1, the cleft regions and internal tunnels of the protein provided potential sites for ligand accommodation and access to binding pockets.
Procyanidin C1 bound to MYBPC3 protein with a binding energy of -6.88 kcal/mol and a Ki of 7.01 µM (Figure 4). This ligand was located in cleft region 2 and interacted with key residues, including Tyr25, Asp32, and Glu48. Other tunnels and pores within the protein provide potential pathways for ligand access to the active regions.
Overall docking results indicated that all three procyanidin compounds are capable of binding to the MYBPC3 protein, and interactions with specific residues in cleft regions and protein tunnels contribute to the stability of the binding. These interactions may provide a basis for investigating the biological effects of OPCs on MYBPC3 function and pathways associated with HCM.
5. Discussion
The PAL gene plays a key role in the biosynthetic pathway of phenolics and OPCs and is of high importance in plant defense responses (16). The sequence extracted from P. persica exhibits stable and balanced physicochemical properties, enabling functional and structural analyses. The molecular weight and pI of peach PAL are consistent with values reported in other plant species. The low Instability Index and long predicted half-life indicate that the protein remains stable under cellular conditions. Additionally, the negative GRAVY value reflects the protein’s affinity for aqueous environments, which is essential for its activity in the cytoplasm (9). The three-dimensional model of PAL showed that the protein functions as a homo-tetramer, and the high quality of the model (GMQE, QMEAN, and Ramachandran favored residues) confirms its suitability for functional analyses and docking with bioactive compounds (7). This information provides an important foundation for exploring PAL interactions with OPCs and understanding its role in phenolic biosynthetic pathways.
The selection and optimization of three-dimensional structures of OPCs play a critical role in analyzing molecular interactions with the PAL protein in peach (17-19). Procyanidins B1 and B2, with directly obtained stable 3D structures, and procyanidin C1, through precise 2D-to-3D conversion, serve as suitable models for simulation and docking studies. Energy optimization using the MMFF94 algorithm adjusted the spatial geometry of the compounds to minimize potential energy while preserving active phenolic groups and B-type inter-unit linkages (19). Optimized 3D molecular structures enable accurate prediction of protein interactions, providing a reliable foundation for bioinformatics and molecular simulation studies.
Bioinformatic analysis of the MYBPC3 gene and protein revealed that this protein plays a crucial role in cardiac function and sarcomere stability (20). The diversity in secondary structure, comprising α-helices, β-sheets, and random coils, highlights the protein’s flexibility and its ability to interact with other sarcomeric proteins. The predicted three-dimensional model enables the identification of functional domains and the analysis of potential ligand-binding sites (7). This information is highly valuable for investigating the effects of OPCs on MYBPC3 function. Given the protein’s essential role in regulating muscle contraction and maintaining sarcomere structural integrity, structural analysis provides a basis for designing small-molecule drugs and intervention strategies for diseases associated with MYBPC3 dysfunction (21). Ultimately, the use of the AlphaFold model allows researchers to predict potential molecular interactions, structural changes, and impacts on cardiac function, providing a foundation for future studies aimed at modulating MYBPC3 activity (7).
The interactions of three OPCs — procyanidin B1, procyanidin B2, and procyanidin C1 — with the MYBPC3 protein were investigated via molecular docking using AutoDock 4.2 (15). The protein and ligand structures were converted to PDBQT format, and the target region included the active sites and potential binding pockets of the protein. Procyanidin B1 exhibited a binding energy of -7.39 kcal/mol and a Ki of 3.78 µM, indicating a relatively strong interaction with MYBPC3. Interaction analysis revealed contacts with amino acid residues Glu48, Phe51, Asp32, and Tyr25 within cleft 2. Cleft, pore, and tunnel analyses indicated that MYBPC3 possesses multiple potential pathways for ligand accommodation, with procyanidin B1 positioned in a partially buried site (22). Procyanidins B2, B1, and C1 bind effectively to MYBPC3 with high affinity, interacting with key residues within protein clefts and tunnels. This stable binding provides a foundation for exploring their biological effects on HCM pathways (23, 24).
5.1. Conclusions
This study used bioinformatics and molecular docking to explore the role of the PAL gene in peach (P. persica) and the effects of OPCs on target proteins. The PAL protein showed stable properties and a homo-tetramer structure, facilitating functional analysis. The OPCs (procyanidin B1, B2, C1) were optimized for molecular interactions. Docking revealed OPCs bind to PAL and human MYBPC3, suggesting potential cardioprotective effects via MYBPC3 interaction. The findings support further research on natural compounds in plant phenolic biosynthesis and cardiac protein regulation, aiding plant genetic engineering and drug design.