1. Background
Coronary artery disease (CAD) is an important cause of morbidity and mortality in cardiac patients. Atherosclerosis, as the underlying pathology of coronary heart disease, is a common multifactorial disorder with both genetic and environmental components (1). Smoking and high blood pressure are the main risk factors for cardiovascular diseases.
Lately, plasma homocysteine has been introduced as a risk factor for CAD (1, 2). It is reported that mutations in 100 genes result in the development of CAD (3). Mutations in genes affecting plasma homocysteine level may be associated with CAD. The association of CAD with single nucleotide polymorphisms (SNPs) in 2 important genes, ie, 10-methylene tetrahydrofolate reductase (MTHFR) and methyltetrahydrofolate-homocysteine methyltransferase (MTR), has been reported in the literature. Several studies on different populations have demonstrated a significant association between polymorphisms, rs1801131 and rs1805087, and development of CAD (4, 5); nonetheless, no studies have been performed in Iran.
Homocysteine is an amino acid, produced during methionine metabolism. It has emerged as an independent atherosclerosis risk factor and a predictor of coronary lesions (6-8). Homocysteine has 2 metabolic pathways: cycle of activated methyl groups and transsulfuration pathway to cystathionine and cysteine. The MTHFR enzyme converts homocysteine to methionine (9). Genetic defects in enzymes involved in the metabolism of homocysteine may increase the level of homocysteine (10). A polymorphism in MTHFR gene (A1298C, rs1801131) is associated with decreased enzyme activity (11). Mutations in MTHFR gene can be found in 30% of individuals with increased homocysteine levels (12).
The MTR gene encodes an enzyme, known as methionine synthase or methyltetrahydrofolate-homocysteine methyltransferase. This enzyme is involved in the processing of amino acids, as monomers of proteins. The MTR gene is mapped to chromosome 1q43 and is highly polymorphic (13). Specifically, methionine synthase converts amino acid homocysteine to methionine. To function properly, this enzyme requires methylcobalamin and methionine synthase reductase, encoded by the MTR gene. The MTR gene polymorphism A2756G (rs1805087) modulates homocysteine metabolism in association with MTHFR gene variants (14, 15).
With this background in mind, in this study, the association of rs1805087 and rs1801131 SNPs with risk of CAD was evaluated in Golestan province via tetraprimer amplification refractory mutation system-polymerase chain reaction (tetraprimer ARMS-PCR) method.
2. Methods
2.1. Subjects
In this case-control study, the population included 120 patients with CAD (70 females and 50 males), who were admitted to Amiralmomenin Hospial in Kordkoy, Golestan province, as well as 130 healthy controls (60 females and 70 males), who were selected from the healthy population of Golestan province with respect to the confounding variables. All participants provided written informed consents before the study.
2.2. DNA Extraction
Genomic DNA extraction from peripheral blood was performed, using the salting out procedure (16). In this method, 2-mL whole blood was transferred to a 15-mL tube and resuspended with 4 mL of cell lysis buffer. This step was repeated to obtain a white pellet. The white blood cell (WBC) pellet was transferred to an Eppendorf tube, and 450 µL of WBC lysing solution, 30 µL of 10% sodium dodecyl sulfate (SDS), and 200 µL of proteinase K were added.
After pipetting up and down a few times, the solution was incubated overnight at 37°C. Then, 1 mL of saturated sodium chloride (NaCl) was added to each tube and shaken for 15 seconds, followed by centrifugation at 2500 rpm for 15 minutes. The upper layer, containing DNA, was transferred to another 15-mL tube. Then, 2 volumes of absolute ethanol were added at room temperature, and the tubes were inverted several times until DNA precipitated. The DNA pellet was dissolved in 50 - 100 µL of H2O.
2.3. Genotyping of rs1801131 and rs1805087 Polymorphisms
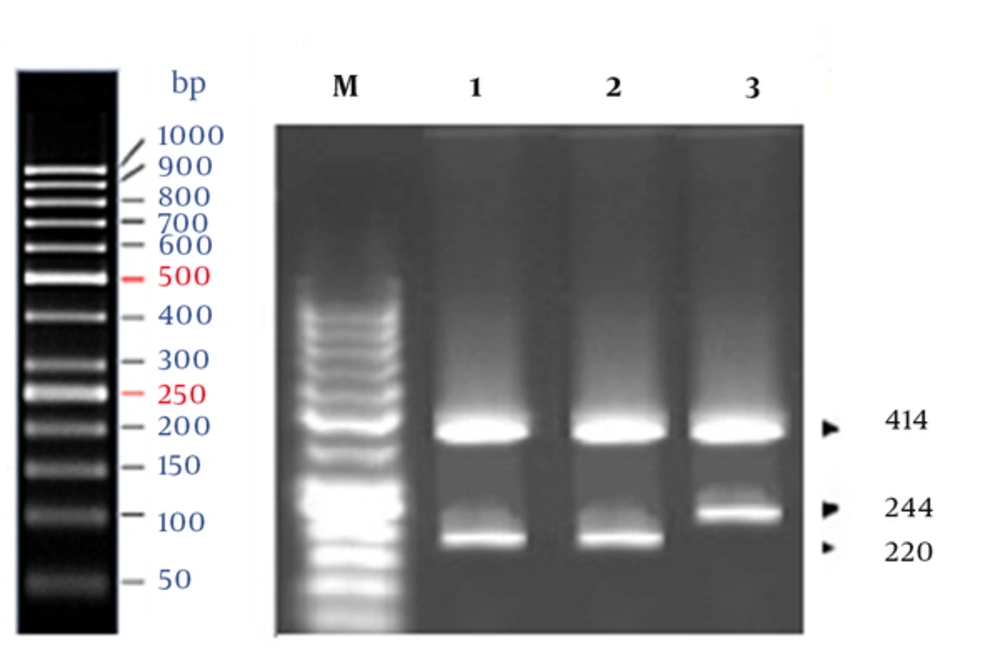
The genotypes for rs1801131 (A/C) in MTHFR gene were specified, using the tetraprimer ARMS-PCR method, according to a study by Masud and colleagues (4). In this method, four primers were used to amplify 3 fragments: 414 bp, 220 bp, and 244 bp (Table 1). Two allele-specific amplicons were generated using 2 pairs of primers: a pair producing an amplicon representing the A allele (220-bp band) and a pair producing an amplicon representing the C allele (244-bp band).
| Genes | Primers | Sequences (5’ .... 3’) |
|---|---|---|
| MTHFR | Outer forward primer | GAGTCAGGGGCAGAATTTACAGGAATG |
| Outer reverse primer | TTCTCCCTTTGCCATGTCCACAG | |
| Inner forward primer | GGGGAGGAGCTGACCAGTGAGGA | |
| Inner reverse primer | AAAGAACGAAGACTTCAAAGACACCTG | |
| MTR | Outer forward primer | GTGTTATCAGCATTGACCATTACTAC |
| Outer reverse primer | GAAGACCTCTGATTTGAACTAGAAGA | |
| Inner forward primer | TGGAAGAATATGAAGATATTAGACATGA | |
| Inner reverse primer | ACTTACCTTGAGAGACTCATAATTGC |
A 414-bp amplicon was generated using the outer primers. Genotyping for rs1805087 (A/G) site in the MTR gene was also performed via tetraprimer ARMS-PCR assay (4). Therefore, two outer primers, producing 328-bp, 203-bp, and 179-bp fragments, were used (Table 1). The 203-bp amplicon represented the A allele, whereas the 179-bp amplicon corresponded to the G allele.
Each PCR reaction contained 100 ng of template DNA, 0.5 mM of each primer for rs1801131 (Sinaclon, Iran), 0.8 mM of each primer for rs1805087 (Sinaclon, Iran), 0.2 mM of each dATP, dCTP, dGTP, and dTTP (KBC, Iran), 1 mM of MgCl2 (KBC, Iran), 2U Taq polymerase (KBC, Iran), and 2 µL of 10X PCR buffer containing 10 mM of tris-HCl (pH, 8.85), 250 mM of potassium chloride, and 50 mM of ammonium sulfate (KBC, Iran). The PCR products were detected through electrophoresis on 1.5% agarose gel.
2.4. Statistical Analysis
In this case-control study, differences in genotype distributions and allele frequencies were determined in the control and case specimens, using standard Chi square test. Odds ratios (ORs) were also calculated.
3. Results
The results of marker association analysis for rs1801131 SNP in our case-control study are presented in Tables 2 and 3. Also, Figure 1 shows the results of agarose gel electrophoresis of ARMS-PCR-amplified fragments for rs1801131 polymorphism. Genotype frequencies of AA, AC, and CC in the case group were 51%, 34%, and 15%, respectively, while the corresponding frequencies in the controls were 43%, 38%, and 19%, respectively. There was no significant difference in the genotype frequency distribution between the case and control groups (P= 0.7943).
| Genes | Alleles | Cases (%) | Controls (%) | P Value | χ2 | Odds Ratio |
|---|---|---|---|---|---|---|
| MTHFR | A | 148 (61.67) | 178 (68.46) | 0.1115 | 1.592 | 1.34 |
| C | 92 (38.33) | 82 (31.64) |
| Genes | Genotypes | Cases (%) | Controls (%) | P Value | χ2 | Odds ratio |
|---|---|---|---|---|---|---|
| MTHFR | AA | 67 (51.53) | 51 (42.5) | 0.333 | 2.19 | 0.69 |
| AC | 44 (33.84) | 46 (38.33) | ||||
| CC | 19 (14.63) | 23 (19.17) |
The rs1805087 marker in the MTR gene was not a polymorph in the investigated population, and all the samples contained AA genotype (two 328- and 203-bp bands). The results showed no significant difference in A and C allele distributions of rs1801131 between the case and control groups (P > 0.05), and there was no significant difference in genotype frequency distribution between the groups (P > 0.05). Consequently, there was no significant relationship between mutant genotypes (A/C or C/C) and CAD in patients from Golestan province (OR, 0.69).
4. Discussion
The present study is the first report of rs1801131 and rs1805087 polymorphisms in homocysteine pathway genes and their association with CAD in Golestan population. In this study, there was no significant difference in A and C allele distributions of rs1801131 among the controls and patients (P = 0.1115). The results suggest that the presence of C allele does not increase the risk of CAD in the population of Golestan province. These results were consistent with those reported by Masud et al., which indicated that the mutated C allele of rs1801131 is not associated with the increased risk of CAD in a population from Pakistan (OR, 1.02) (4).
On the other hand, the present results are inconsistent with a previous study on CAD in a population from Western Europe (5). Szczeklik proposed that MTHFR 1298 C allele is associated with the risk of CAD. MTHFR is an enzyme, which regulates methylation pathways. Genotype determination for A1298C SNP can improve the predictive value of molecular analysis of MTHFR gene in CAD (5). Overall, A2753G (rs1805087) is the most common mutation in MTR gene. Masud et al. observed that rs1805087 (A/G) was associated with CAD in Pakistan (4). However, genotyping of rs1805087 in the present study demonstrated no variety at this position in the population of Golestan province.
4.1. Conclusion
Altogether, these results demonstrated that allele frequency for each marker is specific to each population, and it is necessary to analyze these known markers in other populations. We conclude that there is no significant relationship between rs1801131 polymorphism and risk of CAD in Golestan population. We also observed homogeneity in A2753G (rs1805087) site in the MTR gene in this population. Further similar studies on a larger sample are needed.