1. Background
Following the confirmation of the first case of Covid-19 in Wuhan, China, in November 2019, the virus has rapidly spread worldwide. Hence, the World Health Organization (WHO) declared the outbreak a global pandemic (1). In general, coronavirus expresses four different structural proteins: envelope (E), membrane (M), nucleocapsid (N), and spike (S). Spike protein is related to binding and entry into the host cell. The spike consists of S1 and S2, which attach to the ACE2 receptor of the target cell with S1 (2, 3). Since the onset of the Covid-19 pandemic, various mutations in different parts of the viral genome have been identified in several countries. Some of these mutations resulted in an increased infection rate or changed the pathogenicity. Tens of thousands of SARS-CoV-2 variants differ in at least one conversion (4). Due to the crucial effects of mutations in all stages of the infection (ie, from prevention to treatment), identification and examination of viral variants are critical health approaches in every country. Some variants are less detectable by the reverse transcription-polymerase chain reaction (RT-PCR) technique. It can also be crucial for future decisions, such as complying with lockdown or implementing health protocols and deciding how to control and deal with the virus. Type and manner of usage of drugs, such as the time and dose of drugs, can vary in different variants (5). Dealing with a virus with other structures is the most critical issue in vaccine design because the previously produced vaccines may not be able to fight the new form of the virus genome. Moreover, the SARS-CoV-2 genome mutations vary in different countries, and one type of vaccine may not be helpful in all countries (6, 7). However, the evaluation and identification of mutated virus strains in each region can effectively aid in disease prevention and enforcing health policies in that country.
2. Objectives
This study aimed to investigate mutation patterns in the critical region of the spike gene in the samples of Iranian patients infected with SARS-CoV-2 during the third and fourth waves of the Covid-19 epidemic.
3. Methods
3.1. Sample Collection
In this study, a total of 38 samples were collected from Covid-19 patients whose infection was confirmed using the real-time PCR technique, of which 18 samples belonged to the third wave and 25 to the fourth wave of the Covid-19 pandemic.
3.2. Ethics Statement
The ethical statement was achieved based on the Institutional Review of the Board.
3.3. Extraction of Viral Nucleic Acid
Viral RNA was extracted from 140 µL of viral transport media (VTM) containing polyester tipped swabs of nasopharyngeal and oropharyngeal samples using RN Virus Kit (ROJE Technologies, Yazd, Iran), according to the manufacturer's protocol and was stored in -700C until performing PCR and sequencing.
3.4. Sequence Collection and Primer Design
As a FASTA file, SARS Corona Virus-2 reference sequences were obtained from the genome database. Primers were designed to amplify the spike gene of SARS-CoV-2by OLIGO 7 primer analysis software and synthesized by Sinaclon Company (Tehran, Iran).
3.5. Performing the Molecular Reaction
The spike gene was amplified using Qiagen one-step RT-PCR master mix based on the following conditions: one µL (10 µmole/μL) of each forward and reverse primers (Table 1) were used for cDNA synthesis and PCR amplification. The RNA was converted to cDNA at 50°C for 60 minutes and subsequently incubated at 95°C for 15 minutes to inactivate the RT enzyme and activate the Taq polymerase. PCR was performed on 45 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 68°C for 60 seconds. The PCR products were analyzed by agarose gel electrophoresis (1%) and stained with gel-red safe stain, respectively. The sequencing of samples was performed using the Sanger method.
| Spike Gene | Primer Position (Wuhan COVID-19) |
|---|---|
| GTGCCCTTTTGGTGAAGTTTTTAACGC (outer-forward primer) | 22616 - 22643 |
| CGCCGAGGAGAATTAGTCTGAGTCTG (outer-reverse primer) | 23634 - 23660 |
| TGCTTGGAACAGGAAGAGAATCAGC (inner-forward primer) | 22664 - 22689 |
| CGCATATACCTGCACCAATGGGTATG (inner-reverse primer) | 23600 - 23626 |
The list of Primers Used in This Study
3.6. Evaluation of Mutation in Spike Gene
The quality of sequences was evaluated by Chromas software (Technelysium Pty Ltd., Australia) and analyzed according to the GISAID software algorithm.
4. Results
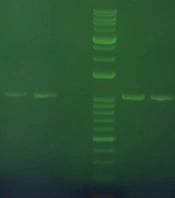
Electrophoresis of spike gene PCR products
The spike PCR products were electrophoresed on agarose gel besides GeneRuler 1 kb DNA Ladder (Figure 1).
4.1. Analysis of Sequenced Samples
Amplicon sequences were read using the Sanger method, and the mutations were evaluated on the dedicated website of Covid-19, the results of which are as follows:
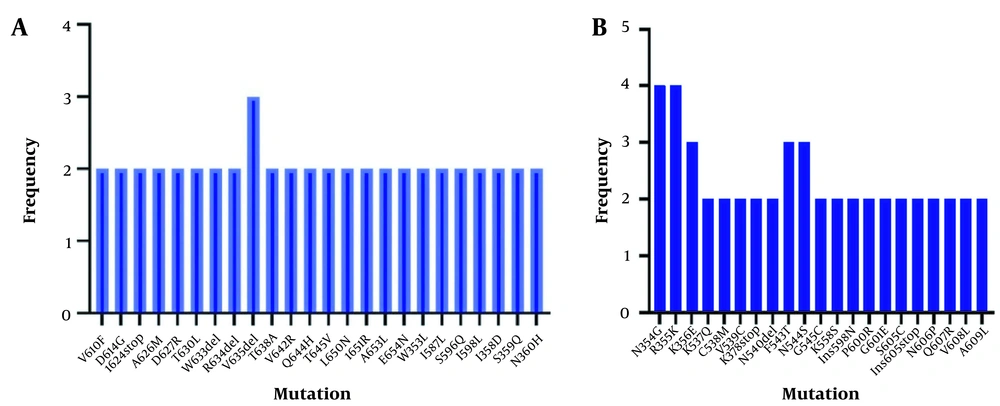
The mutations observed in the third wave of the outbreak are summarized in Figures 2A and 2B.
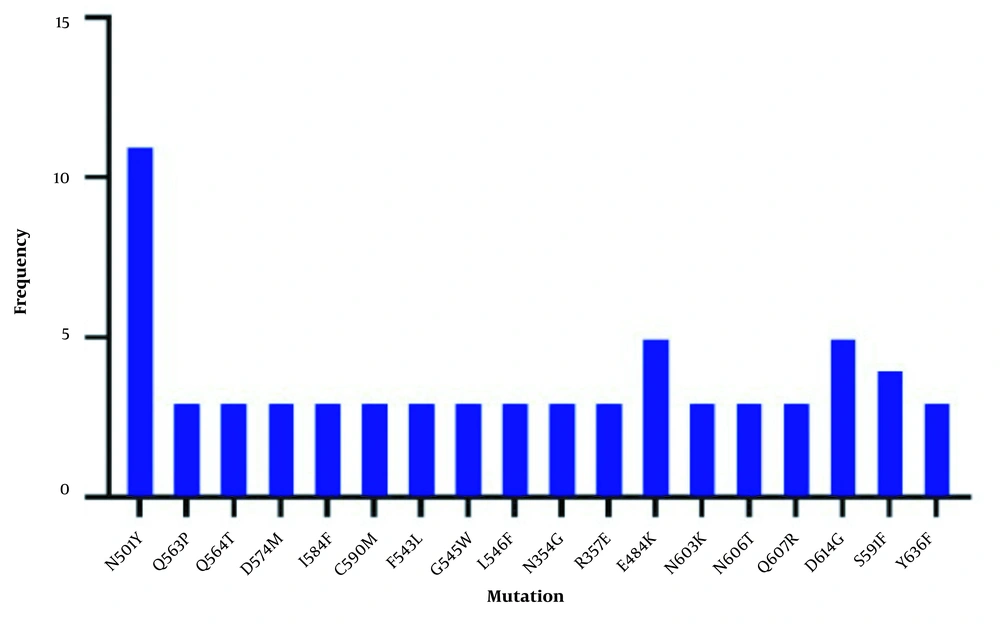
The major mutations observed in the fourth wave of the outbreak are summarized in Figure 3.
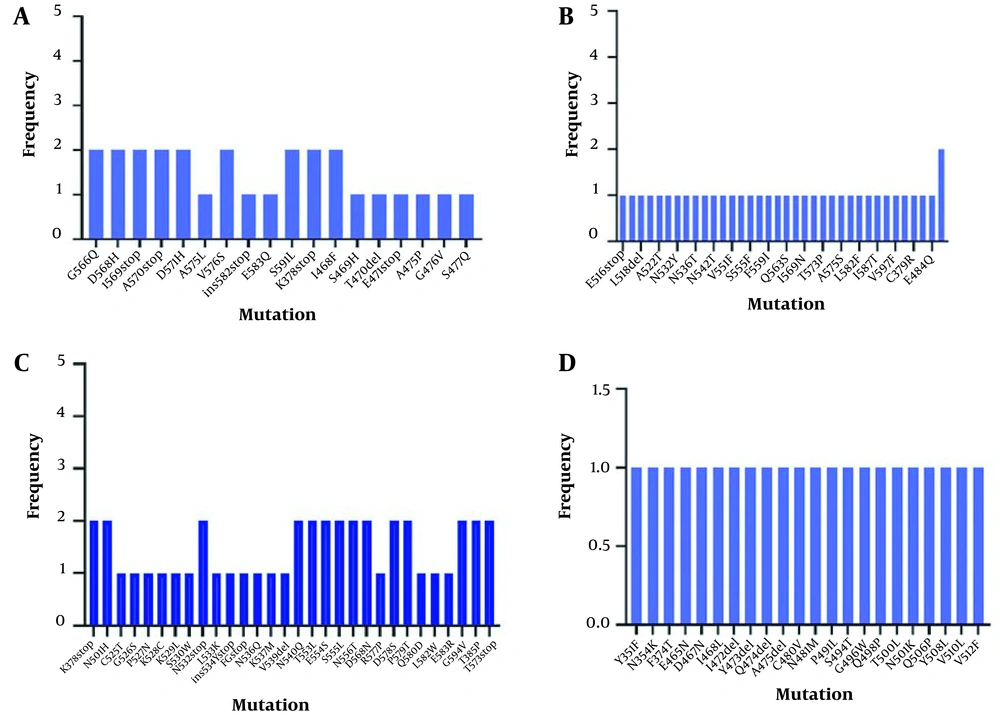
The low-frequency mutations observed in the fourth wave of the Covid-19 outbreak are summarized in Figures 4A, B, C, and D.
5. Discussion
V535deletion, N354G, R355K, K356E, F453T, and F544S were common mutations of the third wave of the Covid-19 epidemic in Iran. The results of sequence analysis of the fourth wave of the Covid-19 epidemic revealed that N501Y, E484K, D614G, and S519F were the most common mutations, and G566Q, D568H, I569 stop, A570stop, V576S, S591L, K378stop, I468F, K378stop, N501H, N532 stop, N540Q, T553L, E554L, N556T, D568T, D578S, P579T, G594V, T385P, and T573stop were significant mutations with lower frequency. The mean value of the real-time PCR cycle threshold (CT) for third and fourth waves' samples were 23 and 16, respectively.
Several significant variants with specific mutations have been reported worldwide, of which essential variants are the B.1.1.7 (first identified in the UK) and the B.1.351 Variants (first identified in South Africa in December 2020 and then discovered in late January 2021 in the United States). Moreover, P.1 Variant was first observed in Brazilian passengers during screening in early January. In addition, the B.1.617.2 variant was first identified in India in December 2020. The B1.1.7Varianthas seventeen different mutations, including N501Y and H69 and V70 deletions. N501Y mutation occurred in the spike protein of the virus and increased the ability to bind to the ACE2 receptor. Omissions are also at positions 69 and 70 of the spike protein (8, 9). According to previous studies, the -L452R mutation in B.1.526.1, B.1.427, and B.1.429 variants is also available in B.1.617 variant.
Moreover, E484K mutation has been reported in B.1.525, P.2, P.1, and B.1.351 variants and some strains like B.1.526 and B.1.1.7. Studies performed in various countries reported a combination of K417N, E484K, and N501Y mutations in the B.1.351 variant (10). There is also a set of K417T, E484K, and N501Y mutations in P.1 that allows these variants to escape from the immune system quickly, and possibly the safety created by the approved vaccines would not be sufficient (11-14). In this study, the mean viral load of the fourth wave samples was much higher than that of the third wave samples. In addition, the same mutations were reported from representatives of the fourth wave. In addition, based on the mutations resulting from sequencing the samples of the fourth wave of the Covid-19 epidemic, it is shown that the dominant strain has mutations similar to the British and South African variants. According to studies conducted in other countries, mutations in the virus genome are associated with an increase in viral titers (15, 16).
5.1. Conclusions
Recently, various variants related to Covid-19 have been reported worldwide. In addition, different mutations have been reported concerning each of the variants, which are related to the proliferation rate and severity of other variants in infected patients. This study showed that SARS-CoV-2 continuously undergoes genetic changes, indicating the necessity of monitoring and evaluating mutations of the dominant circulating variants in controlling the pandemic.