1. Background
Glioblastoma multiform (GBM) is the most frequent, invasive, and undifferentiated kind of primary brain tumor in adults. The median survival time after initial diagnosis is 14 - 15 months (1).
Epidermal growth factor receptor (EGFR) is commonly unregulated in several cancers. The receptor stimulates downstream signals to influence cellular processes (e.g., proliferation, growth, differentiation, and inhibition of apoptosis), and plays a role in both oncogenesis and tumor progression (2).
EGFR inhibitors have therapeutic effects on GBM cases, and the EGFR-Stat5-Fn14 signaling pathway is vulnerable to the population of invasive GBM cells and is probably a suitable target for the treatment of patients. Targeting important effectors in the EGFR–Stat5–Fn14 pathway may decrease GBM tumor dissemination, increase survival chance, and reduce therapeutic resistance (3).
EGFR overexpression has been observed in more than 50% of GBM tumors (4). EGFR activates the signal transducer and activator of transcription (STAT) signaling pathway to promote tumor progression and invasion. STAT is a family of proteins downstream of both receptor and non-receptor tyrosine kinases, including EGFR, which has been shown to contribute to the transcription of many proteins whose positive regulation causes cell cycle progression, aberrant proliferation, and suppression of apoptosis as well as mediates the tumor progression (5).
Direct STAT activation by EGFR binding, as well as indirect STAT activation by Src-mediated EGFR signaling, are some mechanisms used by STAT proteins to mediate intracellular EGFR signaling.
STAT5 is a phosphorylated EGF-dependent protein at unique site and confers new functions on them. Targeting EGFR signaling pathways at numerous levels causes more synergistic therapeutic effects than targeting the upstream receptor alone. Therefore, inhibiting EGFR with regards to oncogenic STATs may serve as a therapeutic strategy for treating cancers by using EGFR signaling re-regulation. So far, no inhibitor has been proven to specifically target STAT, but the essential role of STAT in tumor progression has been reported (5).
It has been revealed that exercise training inhibits cancer in murine models by reducing the circulating concentrations of several hormones, especially insulin-like growth factor-1 (IGF-1), a mitogen causing proliferation by activating Ras-MAPK signaling and preventing apoptosis by activating PI3K-Akt and JAK/STAT pathways (6).
Numerous studies have demonstrated that curcumin prevents the creation of tumor cells and slows down the growth and progression of many cancers (7). By inhibiting the growth factor of vascular endothelial cells and their specific receptor (angiopointin) as well as the angiogenesis of cancer tissue, curcumin reduces the growth of cancer cells and decelerates the activity of telomerase in these cells (8). Although curcumin and exercise training have been found to positively contribute to improving neuro-oncological disorders, data about the potential curative effects of their combination are not sufficient.
2. Objectives
This study aimed to investigate the effects of the combined aerobic-resistance training and nano curcumin on the EGFR/MAPK/STAT5/FN14 pathway genes in the tumor tissue of glioblastoma multiform model rats.
3. Methods
3.1. Animals
A total of 40 healthy male Wistar rats (eight weeks old, 200 - 220 grams) were obtained from the Pasteur Laboratory Animal Breeding and Reproduction Center, Tehran, Iran. They were allowed to eat standard laboratory food and water and were kept for one week in standard conditions regarding temperature, light, humidity, storage place, etc. Then the rats were randomly divided into the control (CON), glioblastoma multiform (GBM), glioblastoma multiform+concurrent training (GBM+CT), glioblastoma multiform+nanocurcumin (GBM+NC), and glioblastoma multiform+nanocurcumin+concurrent training (GBM+NC+CT) groups. All groups included eight rats.
3.2. Tumor Induction
For tumor induction, rats were anesthetized using intraperitoneal injection of xylazine (20 mg/kg) and ketamine (100 mg/kg). Their heads’ hair was shaved, and then the animals were fixed by inserting the rods inside the ears and upper teeth to the stereotaxic device (Stoelting1, model 5504-20019). The bone cap was opened after making an incision in the skin of the skull’s back and removing the periosteum. According to the instructions included in Swanson's Stereotaxic Atlas (1992) (9), the implant position was determined in the following coordinates and marked on the bone as: 2.0 mm laterolateral and 2.0 mm anteroposterior to a depth of 2.5 mm (Figure 1).
Using a Hamilton syringe, 10 microliter of culture medium cells (C6 glioma cell) was implanted in the right frontal cortex. Cells were slowly infused over ten minutes. It was held in position for two more minutes before removing the syringe. To inhibit the administrated solution drawn back into the needle, the syringe was gently lifted until it was completely out of the brain. Then the bone was closed by wax, and the suturing of the skin was performed. Induction of the cancer was confirmed by behavioral tests (Garcia).
3.3. Concurrent Training and Nanocurcumin Consumption
A week before and after the cancer induction, the rats in the training groups were trained to exercise on the treadmill (for 5 - 10 minutes and at a speed of 5 - 10 m/min). Concurrent training program was designed for four weeks. The aerobic training phase was performed for 20 - 35 minutes at 18 m/min on the treadmill, and the resistance phase of the training was performed at the same time in the range of 30 to 100% of the body weight after the rats were tied to weights in the tails and were forced to climb the ladder in three sets with four repetitions (Table 1) (10, 11). All animals were provided with the nanocurcumin supplement by gavage at 100 mg per kg of body weight, five days per week for 28 days (10).
| Exercise Training | Week | Sets and Reps | Intensity | Duration | Frequency | |
|---|---|---|---|---|---|---|
| Concurrent training | Aerobic training | 1 | 18 m/min | 20 min/day | 3 day/week | |
| 2 | 25 min/day | |||||
| 3 | 30 min/day | |||||
| 4 | 35 min/day | |||||
| Resistance training | 1 | 3 sets of 4 reps | 30% BW | 3 min between the sets | 3 day/week | |
| 2 | 3 sets of 4 reps | 50% BW | 3 min between the sets | |||
| 3 | 3 sets of 4 reps | 80% BW | 3 min between the sets | |||
| 4 | 3 sets of 4 reps | 100% BW | 3 min between the sets |
3.4. Measurement of Variables
Real-time PCR reaction was done using SYBR Green PCR master mix and StepOne ABI system to check the expression changes of genes in the studied groups by adopting 2-∆∆ct method. Primers of selected genes were designed using gene runner software (version 6.5) (Table 2). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as the internal control gene. The PCR reaction time and temperature program was carried out as follows: initial denaturation stage at 94°C for three minutes and 35 cycles, annealing stage (temperature of 94°C for 30 seconds), connection stage (temperature of 60°C for 30 seconds), and the amplification or elongation step (temperature of 72°C for 30 seconds) were performed and the final amplification step was set at 72°C for five minutes. The PCR result accuracy and precision were evaluated by using the amplification and melting curves of the PCR product of each gene. Analyzes of gene expression changes were performed using Prism 3 software.
| Gene | Sequence | Product Length | Accession Number |
|---|---|---|---|
| EGFR | Forward: ACAACACCCTGGTCTGGAAG | 193 nt | NM_031507.2 |
| Reverse: GCCCTTCTGGTTGTTGACAT | |||
| MAPK | Forward: CCAACAGGCCTATCTTCCCA | 144 nt | NM_053842.2 |
| Reverse: TTTTGTGCGGGAGAGAAAGC | |||
| STAT5 | Forward: TGGCGAGATCCTGAACAACT | 158 nt | NM_017064.2 |
| Reverse: ACTCAAACAGCACCGTGAAC | |||
| Fn 14 | Forward: AAGGACTGGGCTTAGGGTTC | 153 nt | NM_181086.3 |
| Reverse: TGTGGGGAGCAAGAGATTCC | |||
| GAPDH | Forward: CAAGTTCAAGGGCACAGTCA | ||
| Reverse: CCCCATTTGATGTTAGCGGG |
Information processing in Real-Time PCR, as an important stage, was completed based on standard chart and PCR efficiency evaluation. PCR efficiency is an important factor in relative quantification. To obtain the desired gene to the reference gene ratio, the following formula was used according to the efficiency and difference in CT:
Ratio = E-((ΔCTcase)-(ΔCTcontrol)) ΔCT = CTtarget-CTreference
If the complete efficiency of PCR was desired, the following formula was used:
Ratio = 2-((ΔCTcase)-(ΔCTcontrol))
3.5. Data Analysis
Data were reported as mean ± standard error of means (SEM), were assessed for homogeneity of variance by performing the Levene’s test, and analyzed by using one or two- way ANOVA. Significant difference was checked using a post-hoc test (Tukey) at a statistical significance of α < 0.05.
4. Results
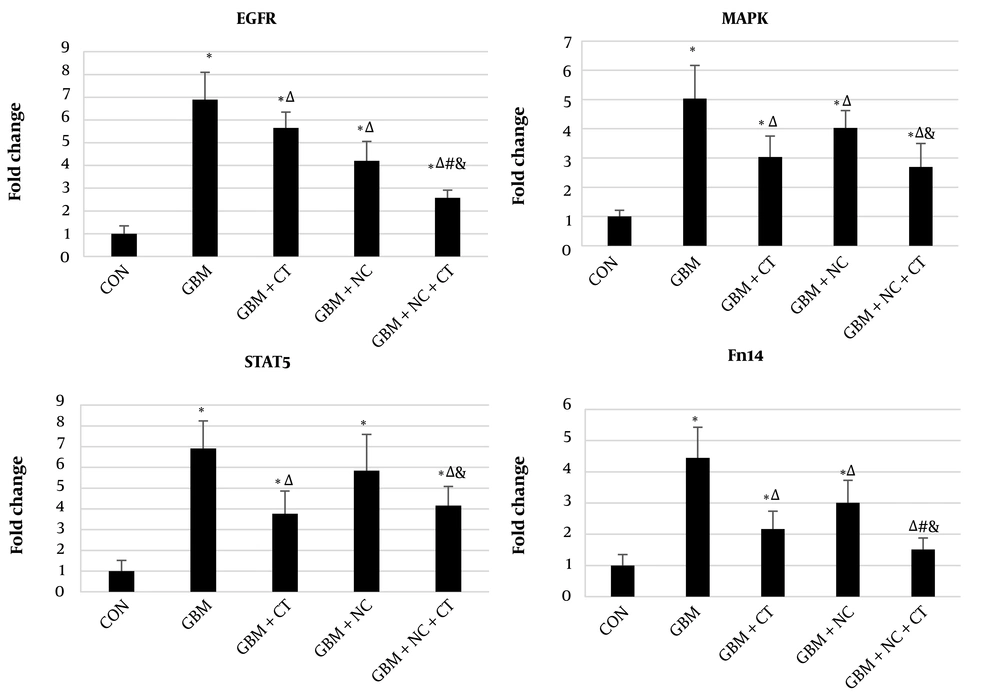
The RT-PCR results demonstrated higher gene expressions of EGFR (739%, P < 0.0001), MAPK (503%, P < 0.0001), STAT5 (834%, P < 0.0001), and Fn14 (445%, P < 0.0001) in the GBM group compared with those in the control group. Concurrent training significantly diminished the expressions of EGFR (18%, P = 0.003), MAPK (39.6%, P < 0.0001), STAT5 (45.5%, P < 0.0001) and Fn14 (51%, P < 0.0001), mRNA compared with GBM group. Nanocurcumin significantly reduced the expressions of EGFR (43%, P < 0.0001) and Fn14 (32.4%, P < 0.0001), mRNA compared with GBM group, but the reductions of STAT5 mRNA (P = 0.47) and MAPK mRNA (P = 0.074) were not significant. Nanocurcumin+Concurrent training decreased the expressions of EGFR (62.6%, P < 0.0001), MAPK (46.2%, P < 0.0001), STAT5 (39.8%, P < 0.0001), and Fn14 (65.9%, P < 0.0001), mRNA compared with GBM group.
EGFR and Fn14 mRNA expression levels in Nanocurcumin+Concurrent training were significantly lower than those in concurrent training group (P ≤ 0.05). Furthermore, significant reduction was recorded for all mRNA expression levels (i.e., 38.6%, P < 0.0001 for EGFR; 32.9%, P = 0.001 for MAPK; 28.8%, P = 0.008 for STAT5; and 49.5%, P < 0.0001 for FN14) in Nanocurcumin+Concurrent training group compared to Nanocurcumin group (Figure 2).
EGFR, MAPK, STAT5 and Fn14 mRNA expression levels in different groups. CON: Control; GBM: Glioblastoma multiforme; GBM+CT: GBM+Concurrent training; GBM+NC: GBM+Nanocurcumin; GBM+NC+CT: GBM+Nanocurcumin+Concurrent training; Data were expressed as means ± SD; 𝑛 = 8 per group; ∗ versus the CON group; Δ versus the GBM group; # versus the GBM+CT group and & versus the GBM+NC group; P ≤ 0.05.
5. Discussion
To our knowledge, the effect of the combination of nanocurcumin consumption and concurrent training on glioblastoma tumors was not known, although the anticancer effects of curcumin and exercise training alone had been determined.
Our study results showed that the gene expression of EGFR/MAPK/STAT5/FN14 pathway in GBM group was significantly higher than that in control group. Ligand binding to EGFR activates some signaling pathways, such as RAS, Src, MAPK, STAT 3/5, PKC, PLCg, and PI3-kinase. Therefore, transphosphorylation and autophosphorylation of the receptors by their tyrosine kinase domains result in the recruitment of downstream effectors and in the stimulation of cell-survival and proliferative signals (12). Researchers, therefore, have invested a great deal of effort to design therapeutic agents to target EGFR. Curcumin is an appropriate option and anticancer drug alone or in combination with other medicines (13). Moreover, exercise has been determined to reduce tumor incidence, growth, and multiplicity in various chemically induced, transplantable, or genetic tumor models (14).
Our findings revealed that nanocurcumin consumption, concurrent training, and both interventions together reduced EGFR/MAPK/STAT5/FN14 pathway genes expression. Bojko et al. reported that combined treatment by low dose of curcumin and low concentration of selective EGFR kinase inhibitors (tyrphostins AG494 and AG1478) dramatically reduced the growth and viability of cultured glioblastoma cells (15). Furthermore, curcumin can block EGFR signaling through the prevention of EGFR tyrosine phosphorylation and suppression of EGFR gene expression mediated by PPAR-γ (16). Jones et al., however, reported contrasting results and observed that 38 days of voluntary wheel running increased the phosphorylated expression of ERK1/2 and did not reduce prostate tumor growth (17).
Suppression of activated EGFR as well as phosphorylated MAPK is associated with maximum tumor growth suppression. Shishodia showed that the blockage of ERK, PI3K/AKT, or JNK signaling had the potential to negatively regulate the expression of PPARγ gene in activated hepatic stellate cells, and reduce the cell apoptosis and growth (18). Curcumin caused a significant inhibition of ERK1/2 phosphorylation, COX-2, and EGFR protein expressions in lung cancer and pancreatic cells, which was associated with reduced survival and increased induction of apoptosis in pancreatic and lung adenocarcinoma cells (19). Curcumin is a strong inhibitor of dendritic cells maturation, which is associated with inhibited activation of MAPKs and nuclear factor-kappaB (NF-κB) as potential targets (20), which were also consistent with our study results.
According to our results, Concurrent training and Nanocurcumin+Concurrent training decreased the expressions of STAT5 and Fn14 mRNA compared with GBM group. Studies have shown that Stat5 can drive cell migration and chemotherapeutic resistance, partly by Fn14 expression up-regulation (3). Microglial cells treated with curcumin showed an elevation in phosphorylation and association with Janus kinase (JAK) 1/2 of Src homology 2 domain-containing protein tyrosine phosphatases (SHP-2), leading to the inhibition of the JAK-STAT inflammatory signaling initiation in activated microglia (21). The JAK1, JAK3, and STAT5 protein levels showed a down-regulation in mice with colitis following curcumin treatment, suggesting that curcumin suppressed JAK-STAT signal activation (22).
Considering the number of growth factors and cytokines regulating EGFR/MAPK/STAT5/FN14 signaling in human skeletal muscle, it is not easy to identify the particular signaling waterfall mechanisms in response to exercise.
The mechanisms responsible for the decrease in ERK1/2 phosphorylation in response to exercise have not received enough research attention so far; however, the possible hypotheses are as follows:
Growth factors (e.g., EGF and IGF-1) and tyrosine kinase receptors (e.g., EGFR or IGF-1R) are known to activate the ERK cascade upon bound (23). Exercise has been shown to reduce systemic levels of IGF-1 in preclinical and clinical breast cancer studies (24), which can inhibit the ERK-MAPK pathway.
Another possible mediator is the modulatory effects of exercise on myokines. Myokines such as irisin, oncostatin, and SPARC have been discovered to inhibit cancer growth in vitro and/or in vivo, (25) and may modulate ERK activation.
Finally, micro-RNAs (miRNA) represent another hypothesized pathway through which exercise counteracts ERK activation. Several miRNAs (e.g., miR-20a, miR-874, and miR-133a) have been indicated to regulate ERK phosphorylation (26). Exercise can modulate miRNA expression in the muscle (27), blood (28) and tumor (29), which can affect ERK activation in cancer cells.
The JAK/STAT signaling waterfall can function as a potential mediator of exercise-related adaptation in human skeletal muscle (30). In addition, the exercise type and intensity cause various signaling responses from JAK/STAT proteins as well as their activating ligands. Upper IL-6, STAT, and GH pathways increase quickly following the exercise and facilitate muscle hypertrophy. STAT and JAK phosphorylation is observed following the aerobic exercise as well as the rapid, acute, and brief resistance exercise (30).
According to our findings, the simultaneous administration of combined training (endurance-resistance) and nanocurcumin were effective in inhibiting the EGFR/MAPK/STAT5/FN14 genes expression in tumor tissue of glioblastoma multiforme model rats. However, performing the nanocurcumin and combined exercise did not have an interactive effect on the expression of any of the target pathway genes. Seemingly, it was a potential therapeutic strategy which may have contributed to controlling and treating the tumor tissue of glioblastoma multiforme; however, it was recommended that further studies should be conducted to corroborate this finding.