1. Background
Pseudomonas aeruginosa is a significant causative agent of Gram-negative infections, especially in individuals with compromised immune systems (1). It is the most commonly isolated pathogen from patients hospitalized for more than one week and a frequent cause of hospital-acquired infections (2-4). In recent decades, the primary concern regarding P. aeruginosa has been its resistance to antibiotics (5). Numerous factors contribute to the antibiotic resistance of P. aeruginosa, including low permeability of its cell wall, the presence of beta-lactamases and cephalosporins, plasmid-mediated aminoglycoside resistance, efflux systems, alterations in antibiotic target sites, and biofilm formation (6-9).
Biofilms are complex, structured communities of microbes enveloped in a self-produced extracellular matrix that supports their coexistence and defense. P. aeruginosa, an opportunistic pathogen, can cause infections both as planktonic cells and biofilm cells (10, 11). Previous studies have indicated that P. aeruginosa biofilms in the airways of cystic fibrosis patients can prolong treatment duration and, in some instances, even result in death (12-14). The reduced susceptibility of biofilms to antibiotics leads to persistent biofilm-based infections despite antibiotic treatment. Antibiotic resistance in biofilm-producing cells is higher than in planktonic cultures, attributed to various factors or genes (10). Multiple mechanisms have been identified for biofilm resistance to antibiotics in P. aeruginosa, including glucan-mediated sequestration, stress response factors, efflux pump expression levels in biofilms, and the expression of specific antibiotic resistance genes in biofilm-forming cells (15, 16).
Recent studies have pinpointed certain genes linked to antibiotic resistance in P. aeruginosa. Hwang and Yoon found that 64 genes might be closely related to antibiotic resistance in P. aeruginosa (15). Several genes, including ahpC, hcpC, hemN, ccoP2, phzF2, oprG (17), PA1875-1877, tssC1, PA2070, PA5033, and PA0756-0757, showed higher expression levels in P. aeruginosa biofilms compared to planktonic cells (10, 18, 19). Specifically, the PA0756 gene is associated with biofilm formation and significantly influences antibiotic resistance development (20). These genes' overexpression was involved in biofilm-specific antibiotic resistance against antibiotics such as tobramycin, gentamicin, and ciprofloxacin. Zhang et al. found that in the P. aeruginosa PA14 strain, genes PA0756, PA2070, and PA5033 were overexpressed, contributing to biofilm-specific antibiotic resistance (10). The toxA gene of P. aeruginosa is widely used for detection purposes as it's an inherent genetic sequence located on the P. aeruginosa chromosome and controls the production of exotoxin A (21-23), a key virulence factor in clinical infections. Exotoxin A acts similarly to diphtheria toxin, hindering protein synthesis by affecting the polypeptide chain elongation factor 2, which leads to substantial tissue and organ damage (24).
Although the roles of PA0756 and toxA genes have been identified, a comprehensive study on their expression in biofilm-forming versus planktonic strains is lacking. Understanding the genes involved in biofilm formation could aid in developing strategies to control biofilms and enhance the treatment and selection of more effective antibiotics.
2. Objectives
This study aimed to explore the expression of PA0756 and toxA genes in clinical isolates of P. aeruginosa, comparing their expression between biofilm-forming strains and planktonic strains.
3. Methods
3.1. Collection of Clinical Pseudomonas aeruginosa Isolates
We collected 160 P. aeruginosa isolates from various hospitals in Tabriz, including 522 Nezaja, Imam Reza, Sina, and Shahid Madani. The bacteria were isolated from different clinical specimens, such as urine, feces, blood, sputum, ulcers, and respiratory secretions. After being transferred to the laboratory, the isolates were re-identified using standard microbiological methods. These methods included observing the colony shape, performing Gram staining, and testing positive in differential culture media, such as the presence of bluish-green pigment, growth at 42°C, and biochemical assays including Triple Sugar Iron (TSI) agar (Merck KGaA, Darmstadt, Germany), sulfide indole motility (SIM) medium (Merck), and Methyl red/Voges-Proskauer (MR/VP) broth (Merck) (25).
3.2. Antibiotic Sensitivity Testing (Kirby-Bauer Disk Diffusion Method)
The Kirby-Bauer disk diffusion method was used for antibiotic sensitivity testing, employing commercially available antibiotic disks. A standard strain of P. aeruginosa ATCC 27853 served as a control (26). The P. aeruginosa strains identified were tested against several antibiotics, including amikacin (30 µg), cefepime (30 µg), tobramycin (30 µg), ceftazidime (30 µg), gentamicin (30 µg), imipenem (30 µg), colistin (10 µg), ciprofloxacin (5 µg), piperacillin (100 µg), gatifloxacin (5 µg), and polymyxin B (300 U).
3.3. Biofilm Formation Assays
For the biofilm formation assessment, an overnight culture with an optical density equal to 0.5 McFarland standards was prepared. We added 20 µL of these cells to the wells of sterile 96-well flat-bottomed microtiter plates, already containing 180 µL of TSB supplemented with 1% glucose. The plates were incubated for 24 hours at 37°C. Afterward, we removed the contents of the wells and used 200 µL of sterile water to wash away planktonic microorganisms. The biofilms were fixed by filling the wells with methanol for 20 minutes and then air-drying the plates. The remaining adherent cells were stained with 200 µL of 1% crystal violet (V/V) for 15 minutes, and the dye was solubilized with 200 µL of 33% (V/V) acetic acid (27). Positive and negative controls included P. aeruginosa PA01 (biofilm-producing) and E. faecalis (EF) 29212 (non-biofilm-producing), respectively. A microtiter plate reader measured the absorbance at 570 nm. Each sample underwent testing three times. Isolates with OD ≥ 2 were further analyzed for biofilm production using molecular tests (28).
3.4. Protocol Preparation for Bacterial DNA
DNA was extracted using the tissue buffer boiling method. A single bacterial isolate colony was incubated in 20 µL of tissue buffer (0.25% SDS + 0.05 M NaOH) at 95°C for 10 minutes. After incubation, the sample was centrifuged for 1 minute at 13,000 g. We then added 180 µL of Milli-Q water to the centrifuged sample and stored the extracted DNA at -20°C for long-term preservation.
3.5. PCR Assays
The PCR assay confirmed the presence of PA0756 and toxA genes (Table 1). The PCR conditions included an initial denaturation at 94°C for 10 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 45 seconds, extension at 72°C for 90 seconds, and a final extension at 72°C for 10 minutes. The products were visualized and documented using 1.5% agarose gel electrophoresis. All confirmed biofilm-producer isolates were evaluated for the presence of PA0756 and toxA genes by real-time PCR.
| Gene | Forward Primer (5' to 3') | Reverse Primer (5' to 3') |
|---|---|---|
| PA0756 | TGCCAAGGCTGTAGGTGAAC | CGAGCTGCGGATGATCTTCCA |
| toxA | GACAACGCCCTCAGCATCACCAGC | CGCTGGCCCATTCGCTCCAGCGCT |
| rpoD-f | CATCCGCATGATCAACGACA | GATCGATATAGCCGCTGAGG |
Sequence of Primers Used in This Study
3.6. RNA Extraction and Complementary DNA (cDNA) Synthesis
Following the manufacturer's instructions, RNA was extracted from planktonic cultures and colony biofilms using RNX-plus solution (SinaClon Co., Tehran, Iran) and an RNase-free DNase kit (Thermo Fisher Scientific, USA). The bacterial suspension in RNX-plus was vortexed and incubated at room temperature. The aqueous phase containing RNA was carefully extracted, and the supernatant was discarded after isopropanol addition and centrifugation. Pellets were washed with 75% ethanol, air-dried, and dissolved in DEPC-treated water. A NanoDrop spectrophotometer (ND-1000, Wilmington, USA) was used to measure RNA concentration and purity. For cDNA synthesis, three µg of DNA-free RNA was used as per the manufacturer's instructions (Yekta Tajhiz Azma, Tehran, Iran, Cat number: YT4500).
3.7. Quantitative Real-time PCR (qRT-PCR)
The qRT-PCR assay was performed using a real-time PCR machine (Applied Biosystems StepOnePlusTM) with SYBR Premix EX TaqII, TliRNaseH plus (Takara Bio Inc), and specific primers listed in Table 1. The reaction protocol included an initial step at 95°C for 2 minutes and 40 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. The 23S rRNA was utilized as a reference standard.
3.8. Statistical Analysis
Data were analyzed using SPSS software version 21 (SPSS Inc, Chicago, Illinois, United States) through descriptive statistics and t-tests at a 95% significance level.
3.9. Ethical Consideration
This study was approved by the ethical committee of Aja University of Medical Sciences under the approval code IR.AJAUMS.REC.1399.181.
4. Results
We obtained 160 P. aeruginosa isolates from clinical samples, predominantly from urine and wounds (Table 2).
| Clinical Samples (n = 160) | Values a |
|---|---|
| Urine | 55 (34.37) |
| Respiratory secretions | 23 (14.37) |
| Blood | 15 (9.37) |
| Wound | 50 (31.25) |
| Burn | 7 (4.37) |
| Sputum | 5 (3.12) |
| Cerebrospinal fluid | 5 (3.12) |
Distribution of Pseudomonas aeruginosa in Clinical Samples
Table 3 presents the biofilm formation capabilities of P. aeruginosa isolates utilizing the microtiter plate method. Among the isolates, 87% (139 isolates) produced biofilms: 69% formed strong biofilms, 11% medium-strength biofilms, and 7% weak biofilms. Additionally, 21 isolates (13%) did not produce biofilms.
| Total Biofilm Formation 87% (n = 139) | Biofilm Negative (n = 21) | |||
|---|---|---|---|---|
| Status | Strong Biofilms | Medium Biofilms | Weak Biofilms | Biofilm Negative |
| Values, % | 69 | 11 | 7 | 13 |
The Biofilm Formation of Pseudomonas aeruginosa Isolates
Table 4 illustrates the antibiotic susceptibility based on the type of clinical isolation. The highest resistance observed was to gatifloxacin (82.25%), while the lowest resistance was to colistin (2.5%). Furthermore, 60% of the isolates were resistant to four or more antibiotics, categorizing them as multiple drug-resistant isolates.
| Antibiotic | Wound | Burn | Urine | Sputum | Respiratory | CSF | Blood | Total |
|---|---|---|---|---|---|---|---|---|
| AMK | 72 | 82 | 44.3 | 62 | 59.1 | 100 | 71.2 | 70.8 |
| CPM | 66 | 42 | 48.1 | 82 | 40 | 100 | 78.9 | 65.3 |
| TOP | 66 | 82 | 50 | 82 | 78.1 | 100 | 71.2 | 75.6 |
| CZID | 88 | 62 | 57.7 | 100 | 73.4 | 100 | 71.2 | 74.6 |
| GM | 78 | 82 | 46.2 | 82 | 73.4 | 100 | 71.2 | 76.1 |
| IMI | 58 | 82 | 27 | 82 | 68.6 | 100 | 71.2 | 69.8 |
| CL | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| CIP | 70 | 42 | 50 | 0 | 68.6 | 100 | 71.5 | 57.5 |
| PIP | 72 | 62 | 54.9 | 82 | 68.6 | 100 | 71.2 | 72.9 |
| GAT | 76 | 62 | 74 | 100 | 92.4 | 100 | 71.2 | 82.2 |
| PMB | 3.6 | 0 | 2.7 | 0 | 2.5 | 0 | 3 | 3 |
Antibiotic Susceptibility Pattern of Pseudomonas aeruginosa Isolates a
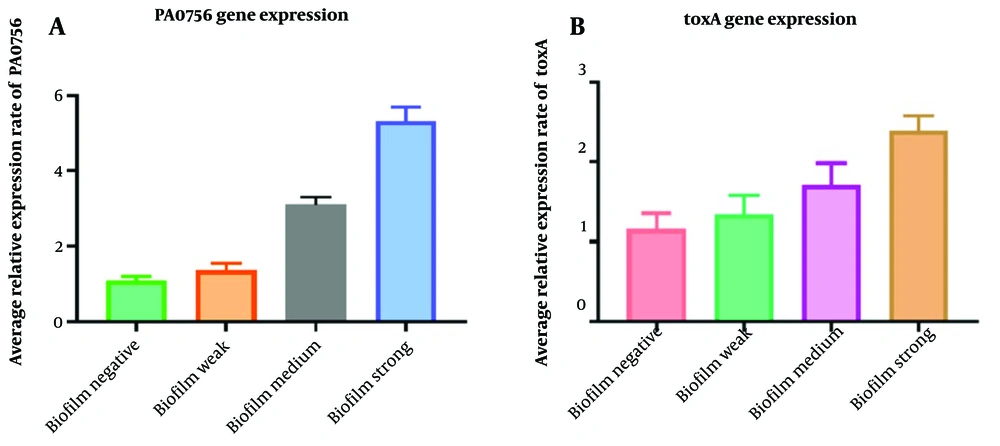
Figure 1 contrasts the expression of the toxA and PA0756 genes in biofilm-producing and planktonic isolates. The expression of the toxA gene was 1.2-fold higher in weak biofilm, 1.7-fold in moderate biofilm, and 2.5-fold higher in strong biofilm compared to planktonic isolates (P = 0.041). Similarly, the expression of the PA0756 gene was elevated in biofilm strains, showing a 1.2-fold, 3.1-fold, and 5.6-fold increase in weak, medium, and strong biofilm strains, respectively, in comparison to planktonic strains (P = 0.012).
5. Discussion
The role of the PA0756 and toxA genes in P. aeruginosa has been the subject of various studies. In our investigation, we analyzed the expression of these genes in clinical infections of P. aeruginosa and compared their expression between biofilm-forming and planktonic strains. We observed a high resistance rate in P. aeruginosa to gatifloxacin, piperacillin, gentamicin, and ceftazidime (82.25%, 72.97%, 76.12%, and 74.63%, respectively), in contrast to a lower resistance rate to colistin and polymyxin B (2.5% and 3%, respectively). These findings are consistent with several studies (29-31). Saderi et al. reported resistance rates as follows: Amikacin (73%), gentamicin (86%), and ceftazidime (73%) (29). Bouza et al. found the highest resistance to gentamicin (72%), tobramycin (69.2%), and amikacin (42%) (30). Franco et al. noted that 34.5% of 238 P. aeruginosa strains were resistant to imipenem, whereas our study showed a resistance rate of 69.84% to imipenem, significantly higher (31). Moreover, our research revealed that of 160 P. aeruginosa isolates, 139 produced biofilms, with most biofilm-forming strains exhibiting resistance to gatifloxacin, piperacillin, gentamicin, and ceftazidime. According to our results, most tested antibiotics displayed high resistance rates, except for colistin and polymyxin B, suggesting these antibiotics may be effective against P. aeruginosa infections.
The increased expression of certain genes, such as PA0756 and the toxA gene, may contribute to biofilm formation in P. aeruginosa isolates. In this study, toxA and PA0756 gene expressions in strong biofilm samples were significantly higher than in planktonic cultures. Similar outcomes have been reported in other research. Darabpour et al. indicated that diminishing toxA expression in P. aeruginosa reduced biofilm formation, potentially due to the toxA gene's impact on biofilm development (32). Zhang et al. identified the upregulation of the PA0756 gene in biofilm cells and its link to biofilm-specific antibiotic resistance (10). Our study examined two genes that are more highly expressed in biofilms than in planktonic cultures, suggesting they may employ unique mechanisms to hinder antibiotic penetration into biofilm cells. It is important to recognize that antibiotic resistance in biofilm cells could involve multiple pathways.
Numerous studies have highlighted the role of the toxA gene in strains resistant to multiple drugs, identifying it as a critical virulence factor that hampers protein biosynthesis, promotes cytotoxicity, and facilitates colonization. Resistant microorganisms are capable of thriving in environments containing bactericidal agents due to the expression of genes that confer antibiotic resistance and enable biofilm production (33). Previous research has indicated that genes encoding putative two-component regulatory systems play a role in biofilm formation and influence antibiotic resistance (10).
This study demonstrated a direct relationship between the toxA and PA0756 genes and biofilm formation in antibiotic-resistant P. aeruginosa strains. Furthermore, diminishing the expression of these genes has been linked to reduced bacterial resistance and lower patient mortality rates in Pseudomonas infections, suggesting that these genes could serve as targets for the development of therapies against antibiotic-resistant infections caused by this bacterium.
5.1. Conclusions
In conclusion, our findings suggest that colistin and polymyxin B, which showed the lowest resistance levels, may be effective in treating infections by multidrug-resistant Pseudomonas. Additionally, the toxA and PA0756 genes were found to be more highly expressed in biofilms than in planktonic cultures, indicating their potential utility in combating antibiotic-resistant P. aeruginosa infections.