1. Background
Sleep apnea is defined by the recurrent ceasing of breathing during sleep, affecting ventilation and fragmentation of sleep. Consequent restorative sleep causes excessive sleepiness and tiredness during the daytime and reduced functionality. The apnea-hypopnea index (AHI) has been frequently used in studies to evaluate the severity of sleep apnea or hypopnea based on events per hour (1). In a previous systematic review, the prevalence of moderate and severe obstructive sleep apnea (OSA) based on AHI was estimated to be 6% to 17% of the general adult population, being more prevalent in males (2). However, a great proportion of affected cases remain undiagnosed. It was estimated that 85% of middle-aged women and 92% of middle-aged men with moderate to severe sleep apnea are undiagnosed in the US (3).
Sleep apnea can increase the risk of hypertension and cardiovascular accidents (4). Although sleep apnea is associated with a high burden, there is usually a lack of adequate attention to this phenomenon (5). Additionally, diagnostic tests, such as polysomnography (PSG), portable monitoring, and home sleep apnea testing, are accurate but time-consuming and expensive (6, 7). Polysomnography, the gold standard for sleep apnea diagnosis, involves the recording of various physiological signals collected from electroencephalogram (EEG), electrooculogram (EOG), electromyogram (EMG), electrocardiogram (ECG), and respiratory signals (8).
Due to the large number of mentioned sensors, PSG might be considered uncomfortable, difficult to perform, and unavailable for many (9). Many recent pieces of research aim to introduce easy-to-do and cost-effective methods and algorithms based on artificial intelligence (AI) to detect sleep apnea. Artificial intelligence refers to the ability of computers and systems to perform tasks that are usually considered to need human intelligence and mental work, such as decision-making and pattern recognition (10).
Methods that focus on singular bio-signals are hot topics in the field of the diagnosis of sleep apnea using AI. Among multiple AI methods that have been developed, convolutional neural network (CNN) are gaining popularity. The convolution neural network is one of the most effective and successful methods inspired by the vision system. Convolution neural network was first developed to classify images (11). Additionally, CNN automatically detects the significant features of the data without any human supervision, which is the main advantage of CNN compared to its predecessors (12). After that, some studies adapted the concept for signal classification by employing a one-dimensional CNN (1D-CNN) network. A two-dimensional CNN (2D-CNN) was also used in studies that converted the one-dimensional signal to a two-dimensional input (13). The combination of CNN and other methods is also gaining popularity (14).
2. Objectives
This systematic review aims to assess the currently developed methods for detecting sleep apnea using CNN.
3. Methods
The current systematic review is reported based on the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines. The targeted outcomes were the accuracy, sensitivity, and specificity of CNN-based methods and the type of signals used in the diagnosis of sleep apnea. For this purpose, all of the studies that have developed CNN-based methods for the diagnosis of sleep apnea and have accomplished the performance tests were eligible for inclusion.
3.1. Search Strategy
Three international electronic databases (PubMed, Web of Science [WoS], and Scopus) were searched from 2010 to October 2023. This time interval was applied because, based on a previous systematic review, no studies have been found before 2010 for CNN-based methods.
The following key terms were used for searching international databases: (1) “Sleep apnea” or “sleep apnoea”; (2) “neural network” or “deep artificial neural network” or “deep learning” or “convolutional neural network” or “recurrent neural network” or “reinforcement neural network” (Appendix 1). No limitations were applied to language or type of study in the search or the selection process.
3.2. Eligibility Criteria
This review included studies that provided data on the following subjects: (1) a CNN-based method was introduced; (2) the method was developed for the diagnosis or staging of sleep apnea; (3) the study has provided data on the performance of the method in diagnostic tests.
This study excluded studies in which (1) they used methods other than CNN; (2) the accurate signal used was not defined; (3) the performance of the test (specificity [SP], sensitivity [SN], and accuracy [AC]) was indicated; (4) only sleep stage classification was performed; (5) different types of sleep apnea without comparison to normal were distinguished; and (6) breathing disorders other than sleep apnea was diagnosed.
3.3. Screening, Data Extraction, and Quality Assessment
Two researchers independently screened the titles of all search results and selected the studies for the abstract screening step. After the abstract screening, researchers screened the eligible full texts and extracted the data. A third researcher reviewed the extracted data. Any disagreements were resolved via consultation with a fourth reviewer in each stage.
The following information was extracted from eligible studies: Bibliometric information (name of the first author and year of publication), the database used or the setting in which the study was conducted, recording sensors or signals, window size in seconds, classification type (apnea, hypopnea, and OSA), classifier type, sensitivity or recall (%), specificity (%), and accuracy (%).
3.4. Statistical Analyses
Due to the heterogeneity of the included studies, it was impossible to conduct a meta-analysis. The data on the SP, SN, and AC of different studies were entered into a Microsoft Excel sheet. The plot was created using the R software (version 4.0.5).
4. Results
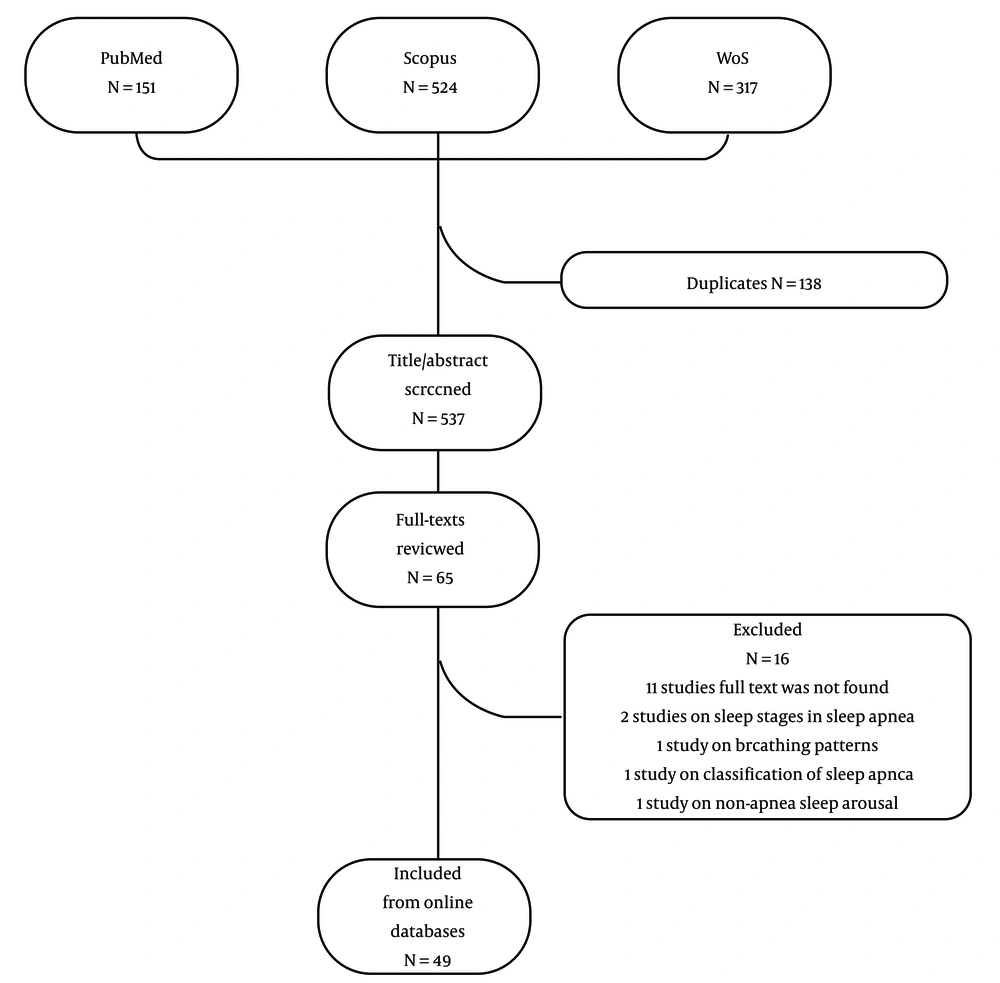
Figure 1 depicts the flow diagram of the study selection process. The titles and abstracts of 659 records retrieved from online databases were screened after the removal of duplicates. Fifty-three studies fulfilled the eligibility criteria for full-text review and were included.
Seventeen studies were excluded as follows: (1) the full text of 11 studies was not found; (2) 2 studies were on sleep stages in sleep apnea; (3) 1 study was on breathing patterns; (4) 1 study was on classification of sleep apnea; (5) 1 study was on non-apnea sleep arousal; (6) 1 study was in a language other than English. Finally, 36 studies were eligible for data extraction.
4.1. Signals for Sleep Apnea Detection
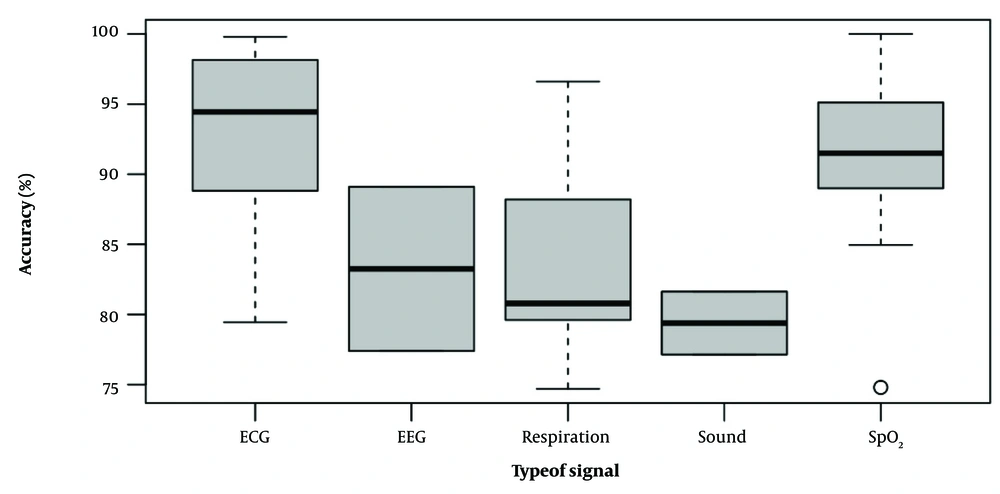
Numerous physiological signals or sensors have been reported to detect sleep apnea based on a CNN classifier. In this systematic review, we found 11 different signals that can be used for CNN-based algorithms to detect sleep apnea, including ECG, blood oxygen saturation (SpO2), sound signals (speech and tracheal sounds), respiratory signals (oronasal airflow and chest and abdomen movements), EEG, combined signals, impulse-radio ultra-wideband (IR-UWB), lateral cephalometric radiographs, and pulse transition time (PTT) (Table 1). Figure 2 shows the boxplots of the accuracy of different signals reported in studies.
| Authors | Year | Database/Setting | Target Group | Recordings | Sensors/Signals | Window Size (s) | Classification Type | Classifier Type | Sensitivity/Recall (%) | Specify (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ECG Signal | |||||||||||
| Zhang et al. (15) | 2021 | AED | Adults | 70 | ECG | 10 | OA/N | LSTM, CNN | 96.1 | 96.2 | 96.1 |
| Shen et al. (16) | 2021 | AED | Adults | 70 | RR-ECG | - | OA/N | MSDA-1DCNN + WLTD | 89.8 | 89.1 | 89.4 |
| Niroshana et al. (17) | 2021 | AED | Adults | 70 | ECG | 60 | OA/N | CNN2D | 92.3 | 92.6 | 92.4 |
| Urtnasan et al. (18) | 2020 | Medical center | Adults | 144 | ECG | 30 | OAS/N | CNN | 99.25 | 98.5 | 99 |
| Thompson et al. (19) | 2020 | AED | Adults | 35 | ECG | 50 | OA/N | CNN1D | 97 | 97.2 | - |
| Sharan et al. (20) | 2020 | AED | Adults | 70 | ECG (HRV) | 60 | A/N | CNN1D | 82.74 | 91.62 | 88.23 |
| Chang et al. (21) | 2020 | AED | Adults | 70 | ECG | 60 | A/N | CNN1D | 81.1 | 92 | 87.9 |
| Per recording | A/N | CNN1D | 95.7 | 100 | 97.1 | ||||||
| Singh and Majumder (22) | 2019 | AED | Adults | 70 | ECG | 60 | OA/N | CNN2D | 90 | 83.82 | 86.22 |
| Liang et al. (23) | 2019 | AED | Adults | 70 | RR-ECG | - | OA/N | LSTM, CNN | 98.97 | 96.94 | 99.8 |
| Erdenebayar et al. (24) | 2019 | Medical center | Adults | 86 | ECG | 10 | AH/N | CNN1D | 99 | 99 | 98.5 |
| CNN2D | 95.9 | ||||||||||
| DNN | 93 | 94 | 93 | ||||||||
| Dey et al. (25) | 2018 | AED | Adults | 35 | ECG | NA | OA/N | CNN1D | 97.82 | 99.2 | 98.91 |
| Banluesombatkul et al. (26) | 2018 | MrOS | Adults | 545 | ECG | 15 | Severe OA (AHI ≥ 30)/N | CNN1D, LSTM, DNN | 77.6 | 80.1 | 79.45 |
| Urtnasan et al. (27) | 2018 | Medical center | Adults | 86 | ECG | 10 | OA/H/N | CNN1D | 87 | 87 | 90.8 |
| Wang et al. (28) | 2018 | AED | Adults | RR-ECG | 35 | OA/N | CNN | 100 | 93 | 97.8 | |
| Urtnasan et al. (29) | 2018 | Medical center | Adults | 82 | ECG | 10 | OA/N | CNN1D | 96 | 96 | 96 |
| Mukherjee et al. (30) | 2021 | AED | Adults | 70 | ECG | 240 | OA/N | CNN, MLP | 84.43 | 88.26 | 85.58 |
| Hu et al. (31) | 2023 | AED | Adults | Raw ECG, RA, RRI, and RRID | NA | OA/N | CNN | - | - | 90.3 | |
| Bahrami and Forouzanfar (32) | 2022 | AED | Adults | 70 | RR-ECG, R-peak | 60 | A/N | CNN | 93.92 | 95.63 | 94.95 |
| Chen et al. (33) | 2022 | AED, UCD | Adults | 95 | ECG | 60 | A/N | CNN | 86.48 | 94.16 | 91.22 |
| Nasifoglu and Erogul (34) | 2021 | ABC, HomePAP | Adults | 292 | ECG | 30 | OA/N | CNN2D | Scalogram images: 83.2; spectrogram images: 81.9 | Scalogram images: 82.2; spectrogram images: 77.2 | Scalogram images: 82.3; spectrogram images: 80.1 |
| Mashrur et al. (35) | 2021 | AED, UCD | Adults | 95 | ECG | 60 | OA/N | CNN | AED: 94.3; UCD: 71.62 | AED: 94.51; UCD: 86.05 | AED: 94.38; UCD: 81.86 |
| SpO2 Signal | |||||||||||
| Mostafa et al. (11) | 2020 | HuGCDN2008 | Adults | 70 | SpO2 | 180 | OA/N | CNN | 74.4 | 94.1 | 89.5 |
| UCD | 25 | 180 | 67.35 | 90.51 | 84.96 | ||||||
| AED | 8 | 300 | 88.58 | 93.67 | 91.5 | ||||||
| Mostafa et al. (36) | 2020 | HuGCDN2008 | Adults | 70 | SpO2 | 300 | A/N | CNN1D | 73.64 | 93.8 | 88.49 |
| HuGCDN2008 | 70 | 180 | AHS | 71.47 | 94.07 | 95.71 | |||||
| AED | 7 | 180 | A/N | 92.36 | 97.08 | 95.14 | |||||
| AED | 7 | 180 | AHS | 92.36 | 97.08 | 100 | |||||
| Vaquerizo-Villar et al. (37) | 2020 | CHAT | Pediatrics | 746 | SpO2 | - | AHS/N | CNN (AHI = 1) | 40 | 98.6 | 74.8 |
| CNN (AHI = 5) | 46 | 98.6 | 90.7 | ||||||||
| CNN (AHI = 10) | 54.2 | 99.6 | 95.1 | ||||||||
| Vaquerizo-Villar et al. (38) | 2019 | CHAT | Pediatrics | 453 | SpO2 | 60 | AH/N | CNN1D | 56.5 | 96.7 | 93.6 |
| Respiratory Signals | |||||||||||
| McCloskey et al. (39) | 2018 | MESA | Adults | 1 507 | Nasal airflow | 30 | A/H/N | CNN1D | 77.6 | - | 77.6 |
| CNN2D | 79.7 | - | 79.8 | ||||||||
| Haidar et al. (40) | 2018 | MESA | Adults | 1 507 | Nasal airflow, abdominal and thoracic plethysmography | 30 | OA/H/N | CNN1D | 83.4 | - | 83.5 |
| Choi et al. (41) | 2018 | Hospital | Adults | 179 | Nasal pressure | 10 | AH/N, G | CNN1D | 81.1 | 98.91 | 96.6 |
| Cen et al. (42) | 2018 | UCD | Adults | 23 | SpO2, oronasal airflow, ribcage, and abdomen movements | 1 | OAH/N | CNN2D | - | - | 79.6 |
| Biswal et al. (43) | 2018 | SHHS | Adults | 5 804 | Airflow, chest and abdomen movements, SpO2 | 1 | AHS | RCNN | - | - | 88.2 |
| MGH | 10 000 | - | - | 88.2 | |||||||
| Haidar et al. (44) | 2020 | MESA | Adults | 1507 | Nasal flow, abdominal and thoracic plethysmography | 30 | A/N | Predictive CNN | 81.73 | 80.63 | 80.78 |
| Haidar et al. (45) | 2017 | MESA | Adults | 100 | Nasal airflow | 30 | OA/N | CNN1D | 754.7 | 74.7 | |
| Wang et al. (46) | 2023 | MESA | Adults | 1 507 | Nasal airflow, abdominal and thoracic expansion | 30 | OA/N | CNN1D, CNN2D, LSTM | 81.73 | 86.59 | 83.90 |
| Sound Signals | |||||||||||
| Simply et al. (47) | 2020 | Medical center, university students | Adults | 398 | Speech signals | 2 | OAH/G | LSTM, CNN | 75 | 79 | 77.14 |
| Luo et al. (48) | 2020 | Hospital | Adults | 132 | Sleep sound | 5 | OA/N | CNN | 69.7 | 70.9 | 81.63 |
| Nakano et al. (49) | 2019 | Hospital | Adults | 1 548 | Tracheal sound | 60 | AH/N | DNN (AHI = 5) | 98 | 76 | |
| DNN (AHI = 15) | 97 | 90 | |||||||||
| DNN (AHI = 30) | 92 | 94 | |||||||||
| EEG Signals | |||||||||||
| Jiang et al. (50) | 2018 | MIT-BIH polysomnographic database | Adults | 18 | EEG | 30 | MSPCNN | 93.1 | 82.9 | 89.1 | |
| Pourbabaee et al. (51) | 2019 | Physionet challenge | Adults | 994 | EEG | 120 | AH/N | DRCNN | - | - | 62 |
| Mahmud et al. (52) | 2020 | EEG dataset | Adults | 25 | EEG | 5 | A/N | FCNN | 83.6 | 77.3 | 77.4 |
| Barnes et al. (53) | 2022 | SHHS2, EEG dataset | Adults | 2 675 | EEG | 30 | A/N | CNN | - | - | 69.9 |
| Combined Signals | |||||||||||
| Alarcon et al. (54) | 2023 | SHHS2 | Adults | 163 | SpO2, heart rate, thoracic respiratory effort, and abdominal-respiratory effort | 30 | OA/N | CNN1D | 82.5 | 86 | 92.1 |
| Jimenez-Garcia et al. (55) | 2022 | CHAT, UofC | Pediatrics | 2 612 | SpO2, airflow signal | 300 | OA/N | CNN | CHAT: 82.4; UofC: 95.2 | CHAT: 99.1; UofC: 93.5 | CHAT: 94.4; UofC: 90.3 |
| Other Signals | |||||||||||
| Arslan Tuncer et al. (56) | 2019 | Hospital | Adults | 100 | PTT | 0.048 | A/N | CNN | 98 | 94.25 | 92.78 |
| Tsuiki et al. (57) | 2021 | Sleep Center | Adults | 1389 | Lateral cephalometric radiographs | Full image | Severe OA (AHI ≥ 30)/N | DCNN | 87 | 82 | - |
| Main region | 88 | 75 | - | ||||||||
| Head only | 71 | 63 | - | ||||||||
| Manual cephalometric analysis | 54 | 80 | - | ||||||||
| Kwon et al. (58) | 2021 | Hospital | Adults | 36 | IR-UWB | 20 | AHS/N | LSTM, CNN | 78.1 | 95.6 | 93 |
| Perez-Macias et al. (59) | 2017 | Hospital | Adults | 30 | Emfit mattress | 30 | A/N | CNN2D | 92 | 96 | 94 |
| Wei et al. (60) | 2023 | Apnea-PPG | Adults | 110 | PPG | Per segment | OA/N | MS-Net | 74.4 | 85.1 | 82 |
| Per recording | 80 | 97.6 | 93.6 | ||||||||
| Jiang et al. (61) | 2023 | Hospital | Adults | 59 | PPG | NA | OA/N | CNN1D | 98.24 | 86.74 | 90.75 |
| He et al. (62) | 2022 | Hospital | Adults | 393 | Craniofacial photos | NA | OA/N | CNN | AHI ≥ 5: 95; AHI ≥ 15: 91 | AHI ≥ 5: 80; AHI ≥ 15: 73 | AHI ≥ 5: 90; AHI ≥ 15: 83.1 |
Abbreviations: A, apnea; H, hypopnea; N, normal; S, severity (based on AHI, OSA is classified as follows: (1) none: AHI < 5 per hour; (2) mild: AHI ≥ 5, but < 15 per hour; (3) moderate: AHI ≥ 15, but < 30 per hour; (4) severe: AHI ≥ 30 per hour); O, obstructive; AHI, apnea-hypopnea index; CNN, convolutional neural network; DCNN, deep convolutional neural network; DNN, deep neural network; RCNN, recurrent convolutional neural network; MSDA-1DCNN, multiscale dilation attention 1-D convolutional neural network; DRCNN, dense recurrent convolutional neural network; WLTD, weighted-loss time-dependent; MSPCNN, multi-scale parallel CNN; AED, apnea-ECG database; CHAT, childhood adenotonsillectomy trial; EEG, electroencephalogram; MESA, multi-ethnic study of atherosclerosis; SHHS, sleep heart health study; MGH, Massachusetts General Hospital; UCD, University College Dublin Sleep Apnea Database; PTT, pulse transition time; RA, R-peak amplitude; RRID, RR interval first-order difference; MS-Net, combined multiscale one-dimensional convolutional block (multi-scale block) with shadow one-dimensional convolutional module (shadow module); MrOS, osteoporotic fractures in men study; PPG, photoplethysmography; ECG, electrocardiogram; SpO2, blood oxygen saturation; IR-UWB, impulse-radio ultra-wideband; HRV, heart rate variability; FCNN, fully convolutional neural network.
4.1.1. Signals Based on Electrocardiogram
In electrocardiography, the electrical activity of the heart is mapped by electrodes connected to the skin. These electrodes detect any minor electrical changes in heart muscle during depolarization and repolarization. Electrocardiogram has been used as a signal for detecting sleep apnea since the early 2000s (9). Sleep apnea can affect the ECG through autonomic nervous system response during sleep (63). Some of these changes include variations in the amplitude of R waves, inter-beat (RR) interval, and baseline fluctuation (64). In this systematic review, of 36 included studies, 16 studies used ECG signals. The majority of these studies have used the apnea-ECG database (AED). This database consists of 70 nighttime ECG recordings (and PSG for some cases) conducted at Philipps University, Marburg, and Germany that are available on the PhysioNet site (65).
Alternatively, Banluesombatkul et al. (26) used the osteoporotic fractures in men study (MrOS) database, which recorded the PSG of 2 991 individuals of 65 years or older at 6 clinical centers (65). Different features of the ECG have been used in this subclass of studies. Heart rate variability (HRV) and RR interval, which is the interval between two successive QRS complexes, are frequently utilized in studies. The RR interval was used by Shen et al. (16), Wang et al. (28), and Liang and Qiao (23). The time window used in ECG-based studies ranged from 10 to 60 seconds, and the sensitivity and specificity ranged from 77.6% to 100% and 80.1% to 99.2%, respectively. The highest accuracy achieved was 99.8% by Liang and Qiao, who used the RR interval (23). Table 2 summarizes the characteristics of datasets used in the included studies.
| Dataset | Setting | Signals Measured | Number of Subjects | Age (y) | Gender | BMI/Weight |
|---|---|---|---|---|---|---|
| AED | University | ECG, PSG for some patients | 70 | 27 - 63 | Both | 53 - 135 kg |
| MrOS | Clinical centers | PSG | 2 991 | +65 | Male | - |
| HuGCDN2008 | University hospital | PSG, diagnosis of sleep apnea by a physician | 70 | 18 - 82 | Both | - |
| CHAT | Multiple clinical centers | PSG | 1 900 | 5 - 10 | Both | Mean: 17.1 kg/m2 |
| MGH | Laboratory | PSG | 10 000 | 42 - 64 | Both | 27 - 36 kg/m2 |
| SHHS | At home | PSG | 5 600 | 55 - 72 | Both | 24.6 - 30.7 kg/m2 |
| UCD | University hospital | SpO2 | 25 | Both | - | |
| MESA | National Sleep Research Resource | PSG, actigraphy | 2 200 | 45 - 84 | Both | |
| EEG dataset | University hospital | EEG | 25 | - | Both | - |
| Physionet Challenge | University hospital | PSG | 1 985 | - | Both | - |
| MIT-BIH polysomnographic database | University hospital | PSG | 18 | 32 - 56 | Both | - |
| SHHS2 | University hospital | PSG | 2 651 | Both | - | |
| Apnea-PPG | University hospital | PSG, PPG | 110 | Mean: 45.25 | Both | |
| ABC | University hospital | PSG | 49 | 18 - 65 | Both | 35 - 45 |
| HomePAP | University hospital | PSG | 243 | > 18 | Both | - |
| UofC | University hospital | PSG | 974 | - | Both |
Abbreviations: BMI, body mass index; AED, apnea-ECG database; MrOS, osteoporotic fractures in men study; CHAT, childhood adenotonsillectomy trial; MGH, Massachusetts General Hospital; SHHS, sleep heart health study; UCD, University College Dublin Sleep Apnea Database; MESA, multi-ethnic study of atherosclerosis; EEG, electroencephalogram; PPG, photoplethysmography; PSG, polysomnography; ECG, electrocardiogram; SpO2, blood oxygen saturation.
4.1.2. Signals Based on Blood Oxygen Saturation
Since decreases in blood oxygen levels are associated with apneic events, the SpO2 signal is a useful and simplified tool to detect these events (66). Blood oxygen saturation could be used as a single signal or in combination with other physiological signals to detect sleep apnea. Databases that are used in this subclass of studies were AED, the HuGCDN2008, childhood adenotonsillectomy trial (CHAT), Massachusetts General Hospital (MGH), sleep laboratory and the sleep heart health study (SHHS), and St. Vincent’s University Hospital/University College Dublin Sleep Apnea Database (UCD). HuGCDN2008 database was collected using the VIASYS Healthcare Inc. (Wilmington, MA, USA) device in the University Hospital of Gran Canaria Dr. Negrin. The database has 70 subjects with PSG recordings in addition to the diagnosis of sleep apnea by a physician (67). The CHAT database is composed of PSG studies of 1 900 children, and apneas and hypopneas were scored using AHI (68, 69). Massachusetts General Hospital and SHHS datasets consist of in-lab (MGH) and at-home (SHHS) PSG recordings as part of routine clinical practice consisting of 10 000 and 5 600 recordings, respectively (65). The UCD database had 25 recordings with SpO2 signals. Four studies have used the SpO2 signal as a single signal (11, 36-38), and two studies have used the SpO2 signal in combination with airflow and respiration signals (42, 43). In two studies conducted by Mostafa et al. (11, 36), different window sizes were tested to get the best accuracy. Overall, the sensitivity and specificity ranged from 40% to 92% and 90.5% to 99.6%, respectively. It seems that the specificity of the SpO2 signal in sleep apnea screening is higher than its sensitivity. Mostafa et al. reached 100% accuracy using the CCN1D classifier in their study (11).
4.1.3. Signals Based on Respiration
Oronasal airflow and respiratory movements are the most direct indicators of breathing disorders and were used by six studies (39-45) to detect sleep apnea. Three studies used nasal airflow as the single signal, and the remaining used a combination of respiratory signals.
The databases used were the multi-ethnic study of atherosclerosis (MESA), MGH, and SHHS datasets. The MESA dataset was collected by the National Sleep Research Resource (NSRR), which includes PSG recordings for 2 200 participants conducted between 2010 and 2011 (65). Each patient was labeled as normal, obstructive apnea, or hypopnea by an expert. By using respiratory signals for the detection of sleep apnea, the obtained accuracy ranged from 74.7% to 96.6%. Choi et al. achieved the highest accuracy in their study, using nasal airflow signal and CNN1D classifier (41).
4.1.4. Signals Based on Sound
The breathing process produces characteristic sounds that can be used to detect sleep apnea (70). This principle was used in three studies. In these studies, tracheal sound, sleep sound, and speech signals were used (47-49). In this model, a CNN is trained to distinguish respiration sounds from environmental noise, and then the trained model is transferred to recognize respiration sounds in sleep apnea. All three studies were conducted in hospital and medical center settings. The sensitivity ranged from 69.7% to 98%, and the specificity ranged from 70.9% to 79%. The best-achieved accuracy in this subclass was 81.63% by Luo et al. (48).
4.1.5. Signals Based on Electroencephalogram
Sleep arousal due to sleep apnea can be detected via EEG, which represents sleep arousal as an abrupt shift in EEG waves (71). Three studies have used this signal, and the methods were fully convolutional neural network (FCNN), dense recurrent convolutional neural network (DRCNN), and multi-scale parallel CNN (MSPCNN) as the classifier, respectively (50-52). The databases used in these studies were the EEG dataset of St. Vincent’s University Hospital, Physionet Challenge, and MIT-BIH polysomnographic database in that order. The EEG dataset of St. Vincent’s University Hospital contains polysomnograms and some other bio-signals of 25 patients. The Physionet challenge is part of the Physionet/computing in cardiology challenge 2018 and includes PSG data from 1 985 subjects who were monitored at the MGH for the diagnosis of sleep disorders (72). The MIT-BIH polysomnographic database, freely available on PhysioNet, includes 18 whole-night PSG recordings (65). The highest accuracy was 89.1%, achieved by Jiang et al. In this study, a sleep apnea detection framework combined with time-frequency analysis for EEG signals and an MSPCNN to learn implicit patterns from time-frequency images were used (50).
4.1.6. Other Signals
Unobtrusive electromechanical film transducer (Emfit) mattress sensor, PTT, IR-UWB, and lateral cephalometric radiographs were used by four studies to detect sleep (56-59). Tsuiki et al. analyzed different parts of lateral cephalometric radiographs to achieve the best performance, and the full image got the best results (57). In this subgroup, sensitivity ranged from 78.1% to 98%, and the specificity was in the range of 82% to 96%. The highest performance was achieved by Arslan with 92.8% accuracy (56).
5. Discussion
In this systematic review, we found different sensors and signals based on CNN algorithms that have been used to detect sleep apnea. Electrocardiogram, SpO2, and respiratory signals were the most utilized signals to detect sleep apnea. Electrocardiogram was frequently used as the single source sensor and was the most studied signal in studies. However, to detect sleep apnea from respiration, it was more common to combine multiple signals. The CNN1D classifier was the most frequently used classifier among various CNN subclasses. The highest accuracy (100%) was reported by Mostafa, using the SpO2 signal and CNN1D classifier. In this study, the global classification of OSA was also reported (11). The 99.8% accuracy was also achieved by Liang (23) using the RR-interval signal in ECG and a combination of CNN and LSTM classifiers. The maximum sensitivity and specificity were achieved by Erdenebayar et al. (99% each) using ECG signals and CNN1D classifier (24).
Studies that used ECG signals accomplished the highest performance in diagnostic tests. However, the majority of the studies that have used ECG signals were tested in public datasets, which are probably cleaner than ECGs in hospital and medical center settings. This could increase the performance of the algorithms in diagnostic tests (30-35, 73). Although sleep apnea is primarily a breathing disorder, respiratory and sound signals were not as successful as ECG and SpO2 in diagnostic tests. In some parts, it is because the algorithms based on sound and respiratory signals are more susceptible to noises caused by environmental and cardiac sounds (74). The AED database was the only database in the current study whose different features (i.e., ECG, SpO2, and respiratory signals) have been used for detecting sleep apnea in different studies. Merely considering this database, there was no superiority of a signal in detecting sleep apnea over other signals. Additionally, the sleep recordings in the AED database are annotated per minute for detecting sleep apnea events (75); however, most of the databases have annotations per second/sample, which follow the rules of the American Academy of Sleep Medicine (AASM) to annotate sleep recordings (76). As respiratory events can be grouped in clusters, and one sleep minute can contain more than one apnea/hypopnea, this could affect the generalization of the results obtained in AED to other datasets and clinical settings.
Using a combination of respiratory signals did not improve the performance of the algorithms in diagnostic tests in comparison with singular SpO2 signals, as usually one of the signals dominates the algorithm. Further studies are needed in this field to optimize the algorithms that use multiple respiratory signals to detect sleep apnea. However, algorithms that use fewer signals or a singular signal are preferred for their simplicity and ease of implementation (77).
A variety of signals were combined in several studies to detect sleep apnea. For instance, for the diagnosis of obstructive apnea, Alarcon et al. combined SpO2, heart rate, thoracic respiratory effort, and abdominal-respiratory effort; nevertheless, Jimenez-Garcia et al. combined SpO2 and airflow signal (54, 55). Since the test is closer to a full PSG by increasing the received signals, it is evident that the use of combined signals could increase the sensitivity and specificity of diagnosis. However, the use of combined signals might increase the complexity and cost of the diagnosis and the susceptibility to noise and artifacts. Therefore, the trade-off between the performance and the feasibility of using combined signals should be considered (54, 78).
The convolutional neural network has recently been utilized to diagnose sleep apnea using photoplethysmography (PPG). By examining the changes in the PPG signal that represent the respiratory and cardiac activities, PPG can be utilized to identify sleep apnea (60, 61). To determine the stages of sleep and the arousal events that occur during sleep, PPG can also record the autonomic nervous system’s modulation. Photoplethysmography is noninvasive, simple to use, and inexpensive compared to other sleep apnea diagnostic techniques. Photoplethysmography, however, has numerous disadvantages, including a sensitivity to noise, signal quality, and motion artifacts. Consequently, it is necessary to evaluate and compare PPG-based sleep apnea detection techniques to the gold standard, PSG (79, 80). Due to the heterogeneity of the results, it is not clear yet whether CNN-based algorithms are more successful in distinguishing apnea and hypopnea together from normal cases, compared to apnea, hypopnea, or normal separately. There was also no significant difference between the accuracy of CNN methods in apnea diagnosis or determining apnea severity. It was also not possible to determine whether the accuracy and other results from online databases are better than in hospital settings. The reason for the latter is that some confounder factors (e.g., classification method, classifier, and window size) are involved, and a deduction could be possible if any other factors were the same when comparing the setting in which the study is conducted.
The majority of the studies used only one database; some of the subjects in these databases were used for the training phase, and the remaining were used for the test phase. The cross-validation between one or more databases was only applied in a few studies. Using one database without cross-validation can limit the generalization of the test results to other populations and settings (14). This study suggests cross-validation with another population or database for future studies. This study also offers to perform a meta-analysis, including running all mentioned methods on the same hardware, to achieve a fair comparison in terms of accuracy and efficiency.
One-dimensional CNN was the most used classifier in the studies. Combining 2 or more classifiers does not necessarily increase the performance of the algorithms. Some algorithms achieved a good performance, although using a less complex model. To obtain an algorithm with the highest performance-to-complexity ratio is of special interest. However, further research in this field is needed to reach a definitive conclusion.
Sleep apnea disrupts normal brain signaling and activity. During episodes, lowered oxygen levels and higher carbon dioxide activate sensors in the brainstem. This triggers the brain’s attempt to restart breathing. Areas involved in controlling breathing, such as the medulla, pons, and amygdala, are impacted (81). Electroencephalogram directly records these brain pattern changes. Convolutional neural networks are suited for EEG analysis since they can automatically learn the spatial and frequency characteristics distorted by sleep apnea (53). Signals measured during sleep studies provide insight into apnea’s neurological effects. Electroencephalogram specifically demonstrates altered wave activity during sleep stages and increased waves with arousal (82). Apneas also affect the timing of signals between EEG electrodes. Convolutional neural network architectures successfully capture abnormalities across electrodes using convolutional and pooling layers, enabling the detection of patterns tied to disrupted breathing (83). Research has achieved over 80% apnea classification accuracy from CNN-processed multi-channel EEG, demonstrating their ability to accurately interpret impacted neural data (84).
Low blood oxygen during apneas influences activity in brain centers controlling breathing. Sensors in the medulla and pons detect oxygen and carbon dioxide changes in blood and brain tissue. This disrupts typical breathing patterns during sleep. Convolutional neural networks learn features in oxygen data linked to these impacts, permitting apnea detection (85). Convolutional neural networks model intricate relationships between oxygen fluctuations and subsequent brain activity.
Abnormal heart rate and rhythms on ECG also reflect neural influences. The autonomic nervous system regulates the heart through pathways originating in the brainstem. Repeated apneas prompt arousals and sympathetic changes are observed as arrhythmias (86). Convolutional neural networks used on ECG can model these control systems and flag irregular rhythms signifying disrupted sleep. This offers insight into apnea’s effects on heart function via the brain-heart connection (21).
Advances in CNN design have boosted AI’s ability to identify apnea-related brain changes. Deeper CNNs with more convolutional filters can model complex EEG/signal relationships (53). Optimizing CNN structures might further aid in unraveling apnea’s neural mechanisms through data analysis (84).
This systematic review provides some insights and suggestions for the future research direction for the diagnosis of sleep apnea using AI methods based on the current literature and the gaps identified in this field. The results of studies in this field can be used to develop home diagnostic tools for sleep apnea, aid clinicians in detecting sleep apnea, and provide a decision support system. Artificial intelligence-based methods demonstrated good feasibility for outpatient application and scalability. However, these methods cannot replace the PSG to date, and PSG is still the gold standard for the diagnosis of sleep apnea in clinical settings.
The current study had some limitations as follows: (1) the studies included in this systematic were heterogeneous and used different sets of signals and classifiers; as a result, conducting a meta-analysis was not possible; (2) this review included studies that used CNN-based algorithms; although the most common and precise type of classifier, it is not the only one; (3) a great number of included studies used online databases, which are public databases from research studies that have met a standardized procedure for recording, annotations, and diagnosis and, therefore, are cleaner than data from clinical settings. As a result, the conclusion regarding the use of AI-based methods for the screening of sleep apnea should be interpreted with caution.
5.1. Conclusions
Algorithms based on a CNN classifier and a singular signal to detect sleep apnea could be recommended as an easy and time-saving approach for aiding clinicians in detecting sleep apnea or home diagnosis. For now, PSG is the gold standard for the diagnosis of sleep disorders, such as sleep-related breathing disorders, in clinical settings. Further studies on the most efficient classifier and cross-validation with clinical settings are needed before AI-based methods can replace the classic approaches.