Although biochemical methods and DNA sequencing have evolved in diagnostic microbiology, they have several limitations in clinical use. The culture method requires a lengthy amount of time to reveal conclusive results, as well as selective and specific media in many circumstances, while numerous clinicians may not have any basic suspicions for microbial diagnosis before laboratory work-up. In contrast, molecular techniques such as polymerase chain reaction cannot differentiate viable or dead organisms and require specific handling. In order to accurately apply molecular methods, we must also be aware of detailed microorganism characteristics. Correct decision-making to choose appropriate molecular methods to assess the probable microorganism is also necessary. Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) is a molecular diagnostic tool with the capacity to analyze nucleic acids, proteins, and sugars. This assay, introduced in 1996, provides very rapid (less than 1 hour) characterization of bacteria at the genus, species, and strain levels (1); MALDI-TOF MS can identify a vast majority of pathogens within hours (2). An increasing number of reports have recognized this tool as a very rapid and reliable method for the detection of numerous types of organisms, such as bacterial and viral species, as well as fungal elements. MALDI-TOF MS works on the basis of characteristic protein profile identification of each microorganism. Elimination of sample manipulation in the different phases of processing and prevention of genome contamination are proposed advantages of MALDI-TOF MS versus molecular-based studies (3). MALDI-TOF MS improves accuracy, reduces costs and time to obtain results, as well as some limitations of other methods. The number of studies using this assay increased slowly until 2011. We searched PubMed with two MESH terms: (“spectrometry, mass, matrix-assisted laser desorption-ionization” [Mesh]) and “microbiology” [Mesh]. For the period since the beginning of 1997 to end of 2006 (approximately 10 years) only 38 results were found, whereas between the beginning of 2007 to end of 2010 50 results and between the beginning of 2011 to the end of 2015, produced 160 results.
Of approximately 31,367 documents (Title contains spectrometry, mass and matrix-assisted laser desorption-ionization) released in Scopus from 2010 to 2014, 37.3% were related to fields of medicine (Figure 1).Figure 1.
Title contains spectrometry, mass, and matrix-assisted laser desorption-ionization (Source Scopus).
1. How MALDI-TOF MS Works
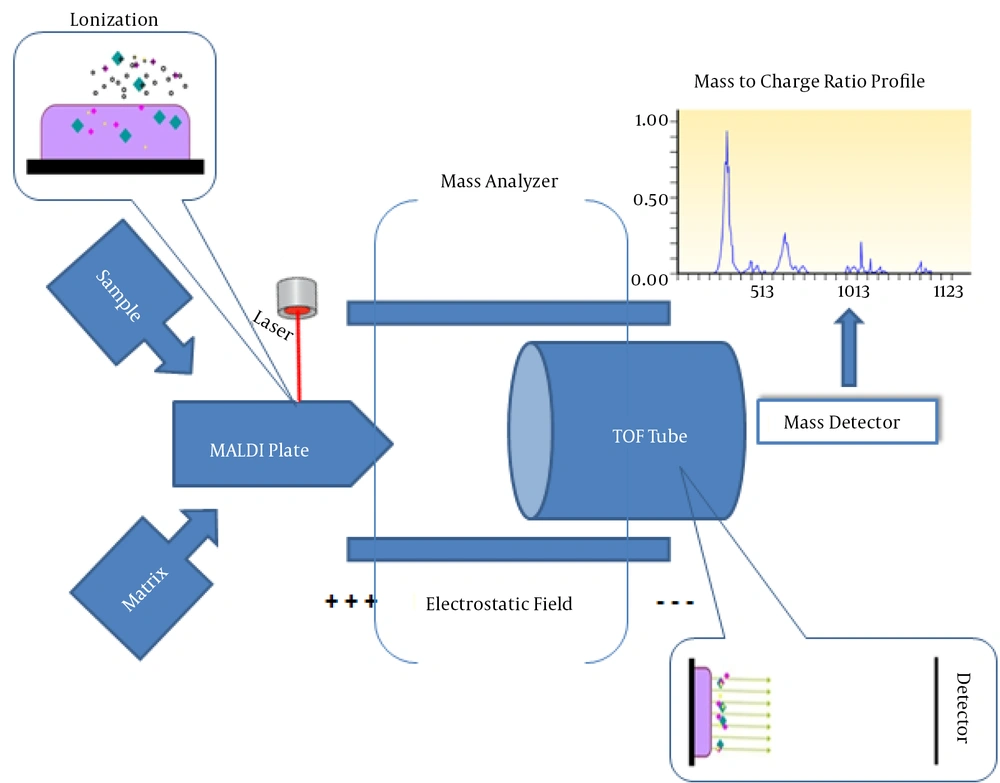
Mass spectrometry measures particles on the basis of their mass-to-charge ratio, which is a physical quantity. Knowing that each particle has a unique mass-to-charge ratio (in terms of predetermined condition), if two particles have a similar such ratio they will be same. A database comprising more than 10 reference spectra (mass-to-charge ratio) per organism species could be valid in identification of a suspected organism. MALDI-TOF MS comprises two components: a MALDI plate and a TOF tube. The MALDI plate is a metal target plate on which the sample (bacteria and yeast from culture media of blood, urine, or cerebrospinal fluid) and matrix are mixed. The matrix is a compound that protects the sample molecules from being destroyed by direct focus of the laser beams and facilitates ionization. After preparation, the mixture is allowed to dry and crystallize. This sample-matrix crystal mixture is then irradiated by laser. After chemical interaction, the sample becomes ionized. Ionization is a critical stage in bacterial identification (for analysis of biomolecules, such as ribosomal protein), after which proteins are analyzed by a mass analyzer (the chamber has an electrostatic field to separate the ions on the basis of their mass-to-charge ratio) to reveal the composition of the sample according to different spectra of mass-to-charge ratios (Figure 2). The TOF tube measures charged ion movement (after separation in the mass analyzer) as it passes through the tube into the detector (which detects and converts ions into a digital output signal) in an apparatus (by comparing the radius of the movement circle with a reference value). Indeed, MALDI is an ionization technique that is used for the identification of bacteria, viruses, and fungi, based on proteomic fingerprints (2, 4).
Sample and matrix are mixed on the MALDI plate, and the dried and crystallized mixture is then irradiated by laser. A chemical interaction sample then becomes ionized. After ionization, proteins are analyzed by a mass analyzer (the chamber has an electrostatic field to separate the ions on the basis of their mass-to-charge ratio) to reveal the composition of the sample. The TOF tube measures charged ion movement as it passes through the tube into the detector.
2. Clinical Use of MALDI-TOF MS
Detection of bacteria isolates from blood cultures in a short time speeds up the identification process. MALDI-TOF MS requires an adequate concentration of the inoculums for successful identification of organisms (5). MALDI-TOF MS has been used with great success to detect bacteria in urine without the need for urine culture. However, it could not accurately identify mixed bacteria present in a urinary specimen (6). Cerebrospinal fluid has also been used for the diagnosis of bacterial meningitis. In addition to Gram stain and culture, use of MALDI-TOF MS could increase sensitivity for early diagnosis. The identification of yeast isolates from positive blood culture had a shortened detection time, according to some reports. MALDI-TOF MS hastens appropriate antifungal therapy by direct identification of yeast species (7). Several recent reports have described the use of MALDI-TOF MS in recognizing antibiotic-resistant bacterial strains. Some of the currently investigated patterns of bacterial resistance include detection of resistance to beta-lactam antibiotics in enteric and non-fermenting gram negative rods (8), carbapenem-resistant Acinetobacter baumannii (9), carbapenem-resistant Klebsiella species (10), carbapenem-resistant Bacteroides fragilis (11), methicillin-resistant Staphylococcus aureus (12), vancomycin intermediate Staphylococcus aureus (13), and vancomycin-resistant Enterococcus (14). Bacterial spectra that have been studied by MALDI-TOF MS contain numerous Gram-positive species, including coagulase-negative staphylococci (15-17), Staphylococcus aureus (18, 19), beta hemolytic streptococci (20, 21), Streptococcus pneumoniae (22), Viridans group streptococci (23), Enterococcus species (24), Bacillus species (25, 26), Listeria (27), Corynebacterium species (28), Arcanobacterium species (29), Nocardia (30), and Mycobacterium species (31). The broad group of Gram-negative bacteria that has been evaluated includes Enterobacteriaceae (32, 33), fastidious Gram-negative bacteria, Brucella (34), Bartonella (35), Francisella (36), Haemophilus (37), Vibrio species (38), Aeromonas (39), Campylobacter (40, 41), Helicobacter (40), Neisseria species (42), Moraxellacatarrhalis (43), and Legionella (44). Propionibacterium (45), Bacteroides species (46), and Clostridium species (47) are anaerobic bacteria that are under investigation by MALDI-TOF MS for prompt and accurate diagnosis. MALDI-TOF MS has also recently been applied for the detection of human polioviruses and enterovirus, and to identify specific viral protein biomarkers in infected cells (3). Among yeasts, Candida (48, 49) and Cryptococcus (50) have been studied. MALDI-TOF MS has shown promising results in the exact diagnosis of mold species, such as Aspergillus and Fusarium (4, 51, 52). MALDI-TOF MS has allowed the identification of the unique spectrum markers of each of the different species. This tool was initially used to evaluate few organisms, but all types of bacteria, viruses, and even fungi, can now could be detected by various types of MALDI-TOF MS. It allows the detection of bacterial macromolecules in complex mixtures without isolation, which can be considered as having distinct superiority over culture-based identification. In addition, MALDI-TOF MS has successfully been used for rapid and accurate identification of difficult-to-identify bacteria from the respiratory tract of people with cystic fibrosis (53). A MALDI-TOF MS-based assay enables the detection of beta-lactamase activity of bacteria within 1 to 3 hours of a positive blood culture. The vast majority of studies regarding the clinical application of MALDI-TOF MS have been published in the United States and Europe, and few Asian countries have begun to start to work with this diagnostic tool. MALDI-TOF MS is now considered as state-of-the-art diagnostic testing for the detection of various microorganisms. Finally, MALDI-TOF MS is not currently FDA-approved for routine diagnostic testing. In the previous edition of our textbook (Feigin, R.D. and Cherry J.D., textbook of pediatric infectious diseases, 2009) and the previous edition of Mandell’s textbook, (Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 2010) MALDI-TOF MS was mentioned only once and three times, respectively. However, this diagnostic modality has been mentioned 10 times in the new edition of the textbook of pediatric infectious diseases (2014) and 40 times in Mandell, Douglas, and Bennett's Principles and Practice of Infectious diseases (2015) (54, 55), and MALDI-TOF mass spectrometry may replace the current traditional methods we use, such as Gram stain, culture, and biochemical tools, in the near future. Although MALDI-TOF mass spectrometry may be an expensive test, it is cheaper than certain conventional routine diagnostic tests, and also provides a definitive diagnosis that may reduce the need for subsequent diagnostic tests and offset the higher upfront costs.
3. Disadvantages and Limitations of MALDI-TOF MS
In a similar manner to other diagnostic tests, MALDI-TOF MS has some limitations. Protein extraction is a highly complex stage in the process of testing, and could decelerate identification of certain organisms, such as Mycobacterium species. The mycobacterial cell wall should be removed for protein extraction with difficult and complex techniques that are sometimes unsuccessful. There is some other identified limitation about this method that may contributing partially in decreasing reported researches after primary trend in its use. MALDI-TOF MS is unable to directly detect organisms on blood samples and needs culture-based amplification step. This may leads to one of the main limitations of MALDI-TOF MS that is decrease identification power when the sample contains low colony count grown on agar or on a blood culture. On the other hand currently MALDI-TOF MS is limited in detection of polymicrobial infections and needs improved identification algorithms. Although specie identification with this method occurs rapidly (<10 min) and will aid clinician to choose appropriate primary antibiotic (or antiviral or antifungal agent) but antibiotic susceptibility testing still is a time consuming step for complete interpretation of primary results. Slow implementation of this method in the clinical microbiology labs may have discouraged researchers toward its use for routine diagnostic practice. MALDI-TOF MS is not currently FDA approved for routine diagnostic testing.
This paper introduces MALDI-TOF MS with the hope that trials conducted by researchers in the field of diagnostic microbiology, with the assistance of laboratory professionals, will begin in Iran.