1. Context
Among recent revolutions in technology, Artificial Intelligence (AI) is one emerging boom that is happening all over the world. Considering supercomputers, facial recognition, robots in surgery, or automated manufacturing, AI has taken over the world. This is the reason why AI futurists and economists tend to call AI the “fourth industrial revolution” (1). Although the mortality rate of the SARS-CoV-2 virus is low, this novel coronavirus (COVID-19) is one of the most infectious diseases affecting our planet in the past decades. This virus was first spotted in Wuhan, China, and has now globally spread to over 190 countries, infecting more than 400,000 people and more than 15,000 deaths (2, 3). Governments and health organizations of the world are constantly working day and night to combat the disease, but it has so far proved to be a major hurdle. Many scientists are seeking the help of AI. Artificial intelligence can aid as an effective tool to study the virus and its capabilities, virulence, and genome. Furthermore, it can help predict the protein structure of the virus and its interaction with other chemical compounds. This can help accelerate the preparation of new antiviral drugs and vaccines (4). Artificial Intelligence and Machine Learning (ML) are two giant effective weapons that we have against this notoriously fast progressive virus. The main aim of our review was to deal with the interaction between viromics and AI/Deep Learning, which might remarkably help find a solution to this pandemic. We also offered our processed data and code after analyzing different articles, which might be used for diagnostic purposes by doctors, researchers, scientists, and virologists who are involved in finding a solution for the SARS-CoV-2 pandemic. This technology is a promising approach in healthcare and can meet the needs of researchers and scientists in antiviral drugs and vaccine development (5, 6). The main objective of this article was to emphasize the importance of AI/Deep learning technologies in the field of healthcare and curbing infectious diseases like COVID-19. Using these tools, researchers can speed up the way of finding a possible cure to COVID-19.
2. Evidence Acquisition
2.1. Data Sources and Research Strategies
A systematic review was conducted on the following databases: MEDLINE/PubMed, SCOPUS, Web of Science, ScienceDirect, and Google Scholar for different studies regarding AI and ML technologies. We used the following combined Medical Subject Headings (MeSH) terms in Google Scholar, such as “Deep Learning”, “Spike Glycoproteins”, “Artificial Intelligence”, “Algorithms”, etc. Finally, the articles were deeply studied, of which key, in-depth information was obtained. The same search process was adopted for other databases.
2.2. Eligibility Criteria
Articles were included on the following basis: (a) majorly published in the English language between 2013 and 2020; (b) accounts of successful trials and experimentations with various AI/Deep Learning technologies such as Tensorflow, Keras, and Python tools; (c) recent WHO and CDC reports of COVID-19; (d) Viral genomic studies; and (e) original and peer-reviewed articles.
The articles were excluded on the following basis: (a) insufficient or no data and (b) articles that did not have a proper study or design approach.
2.3. Tools Employed
NCBI Genome WorkBench was used for processing the sequences, and ClustalX (version 2.1) was used for the alignment of the sequences. NCBI Tree Viewer was used for visualization of the phylogenetic tree. Additionally, python tools were employed in Jupyter Notebook (version to explore the spike glycoprotein structures of betacoronaviruses obtained from different hosts such as bats (Chiroptera), human (Homo sapiens) around the world, rabbits (Oryctolagus cuniculus), brown rats (Rattus norvegicus), and many other possible potential hosts for SARS-Cov-2 updated in NCBI till date. BioPython (version 1.76) was used for importing modules. We used PyMol software for the molecular visualization of the SARS-CoV-2 spike glycoprotein structure.
3. Results
3.1. Artificial Intelligence to Spot Patterns and Changes
The primary way to monitor the spread of infectious diseases is to look for “signs” or “signals”. Artificial intelligence can see these signals in data earlier than humans. In late December of 2019, Li Wenliang warned his fellow colleagues and officials in Wuhan, China, about an upcoming possible SARS-like epidemic that shook the nation in the early 2000s before tragically succumbing to the disease (7). Simultaneously, a computer as AI server named BlueDot also alerted systems regarding an emerging risk of COVID-19 in Hubei province, China. This is not the first kind of disease that this software has alerted about. It has previously detected the outbreaks of 150 different pathogens, including SARS, Zika, Hepatitis A, Measles, etc. (8, 9). Many biotech companies are relying on AI to speed up the way to cure COVID-19. The hope is that it can spot patterns and changes and provide a promising approach for vaccine development. As coronaviruses such as SARS-CoV-2 have tendencies to mutate or even become drug-resistant, the drug developed should be effective for all forms (Broad Spectrum). Many drug companies such as Insilico Medicine Inc., Iktos, Vir Biotechnology Inc., Moderna Therapeutics, and Atomwise are utilizing AI technologies to develop a cure for this disease. Studies estimate that it will probably still take a year or more for the drug to be completely, successfully developed, and introduced to the market. However, in the case of AI, if the tendency of the potential virus is identified easily, the drug development can be achieved at a faster rate (10). Using informative techniques such as AI Neural Networks, Convolutional Neural Networks, Deep Generative Neural Networks, etc., initial potential molecular compounds and viral strains from different places around the world can be analyzed. Although its impact may seem relatively limited at first, this technology has great potential in the field of drug discovery and development (11).
3.2. Viral Host Prediction with Deep Learning
Zoonotic diseases, which involve animal to human transmission of infectious diseases, are current global problems. It could be understood from the recent problem of COVID-19 that wreaked havoc on the world. The first outbreak of SARS-CoV-2 in China can be traced back to Huanan Seafood Market in Wuhan. It is a wet market meaning that meat is sold alongside wild, exotic animals such as civets, bats, pangolins, etc. There could be numerous cases of possible transmission between these animals, either dead/ alive/vectors to humans. In such cases, it is difficult to determine the actual host of the virus -the host may be particularly adapted- and possible origin of the virus (12, 13). To overcome this situation, viral genome structures obtained from different animals, possibly hosting betacoronaviruses such as SARS-CoV-2, are obtained and can be trained with deep neural networks. Various computational techniques that can analyze the DNA and RNA sequences of the virus have been developed. Different architectures have been developed to train neural networks by continuously feeding data (14). Some of the ANN (Artificial Neural Network) based viral host prediction approaches are listed below.
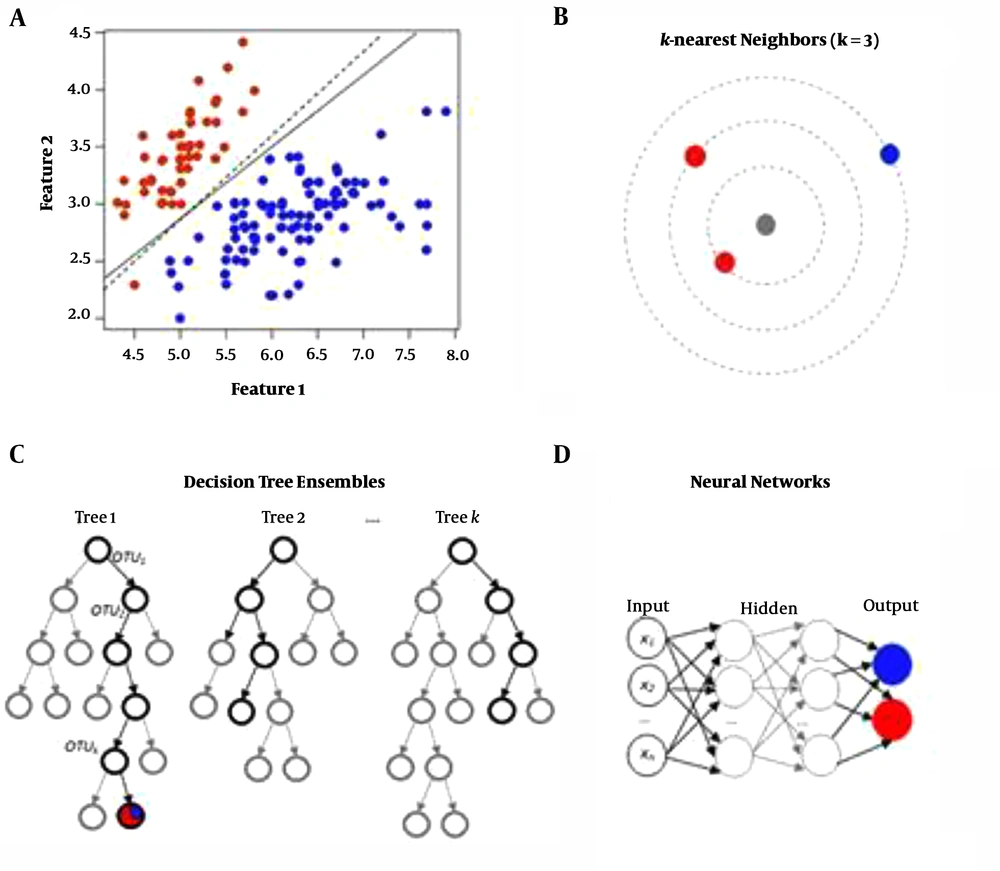
● Truncated Back Propagation Through Time (TBPTT) Algorithm: A viral sequence is processed one timestep at a time (k1 timesteps). It minimizes error for the expected output with a given input (Figure 1).
● Recurrent Neural Networks (RNNs): RNNs can be repeatedly trained to predict viral host traits. By using this method, we can obtain the nearest neighbors and neural networks can be built (Figure 2).
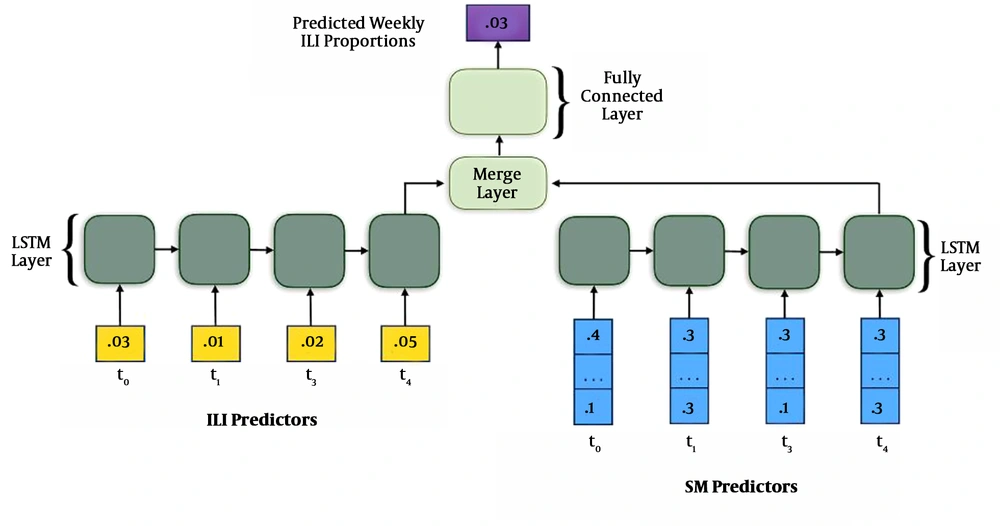
● Convolutional Neural Networks (CNNs): The prediction of virus mutation can be studied using CNNs. This can be used to analyze the course of an outbreak. The below image depicts the forecasting of the dynamics of an influenza-like illness. This same process can be applied to SARS-CoV-2 (15-18) (Figure 3).
3.3. Detecting COVID-19 in X-Ray Images with AI Tools
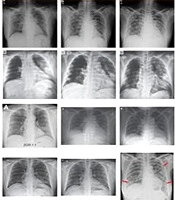
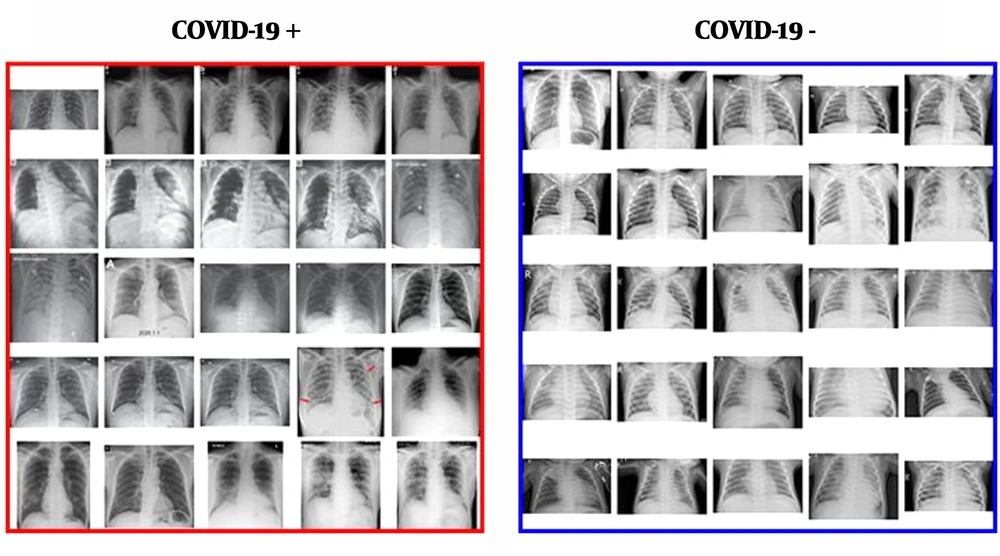
It is possible to detect COVID-19 in X-Ray images of patients using AI tools such as Tensorflow and Keras. Although this method is not a reliable, accurate medical diagnostic method, it still shows how knowledge from computer vision/deep learning can make a big impact on the world of healthcare using AI tools. We just need to obtain the X-ray dataset of patients who have been tested positive for COVID-19. It can be obtained from various open sources. This is followed by sampling of “normal” patients. Finally, a CNN should be trained to detect COVID-19 from the sampled X-Ray images. The below figure shows the dataset of X-Rays of COVID-19 positive and COVID-19 negative patients. These can be trained by neural networks to automatically detect the presence of COVID-19 (19, 20) (Figure 4).
3.4. Exploring SARS-CoV-2 Spike Glycoprotein with Python Tools
Since the discovery of COVID-19, there have been various controversies and debates on its possible origin. The genome of SARS-CoV-2 consists of single, positive-stranded RNA, which is roughly 30K nucleotides long. The genome structure has similarity to other coronaviruses, more specifically the betacoronaviruses. The particular characteristic that makes it similar to other betacoronaviruses is the spike glycoprotein structure. It facilitates the entry of the virus into the cells. In this process, we are going to explore the genome of SARS-CoV-2 from different hosts (21).
3.4.1. Importing Modules
The following modules were imported from BioPython (version 1.76) (22) (Figure 5).
3.4.2. Obtaining Sequences from NCBI
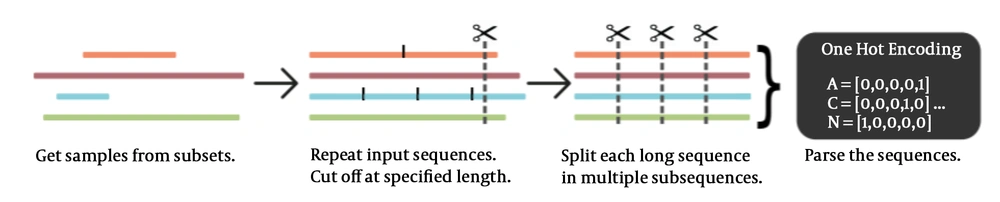
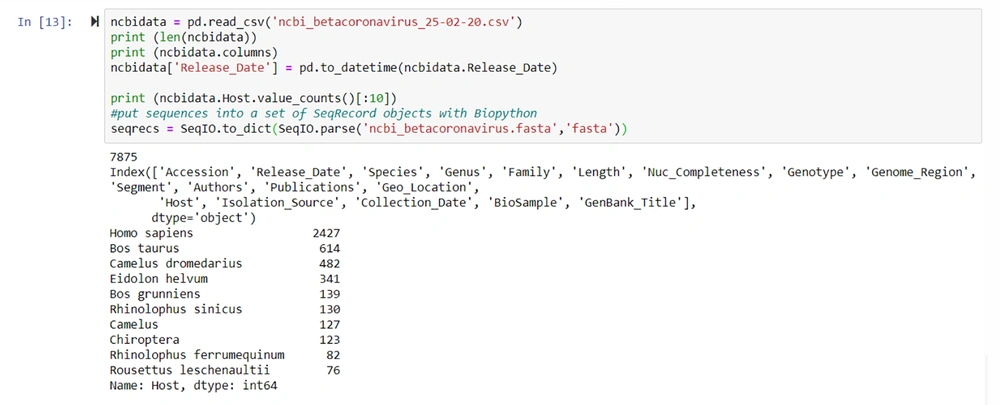
NCBI virus database was used to download the betacoronavirus sequences. The csv and fasta files for the viruses were downloaded. By using the accession number or host, we accessed the subsets (23) (Figure 6).
3.4.3. Getting Subsets of the Sequence
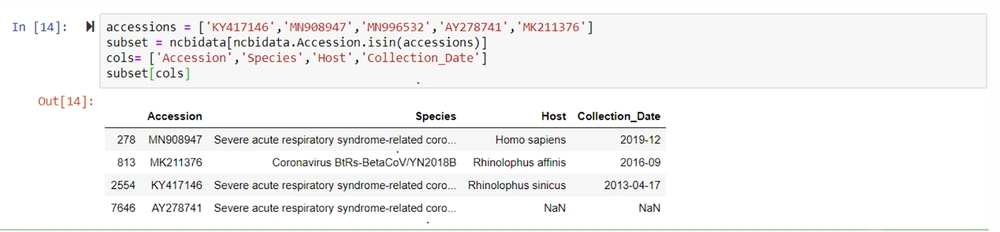
The subsets were obtained after accessing the SARS-CoV-2 betacoronaviruses. It was listed with the species, the host, and the date of collection (24) (Figure 7).
3.4.4. Aligning the Given Sequences
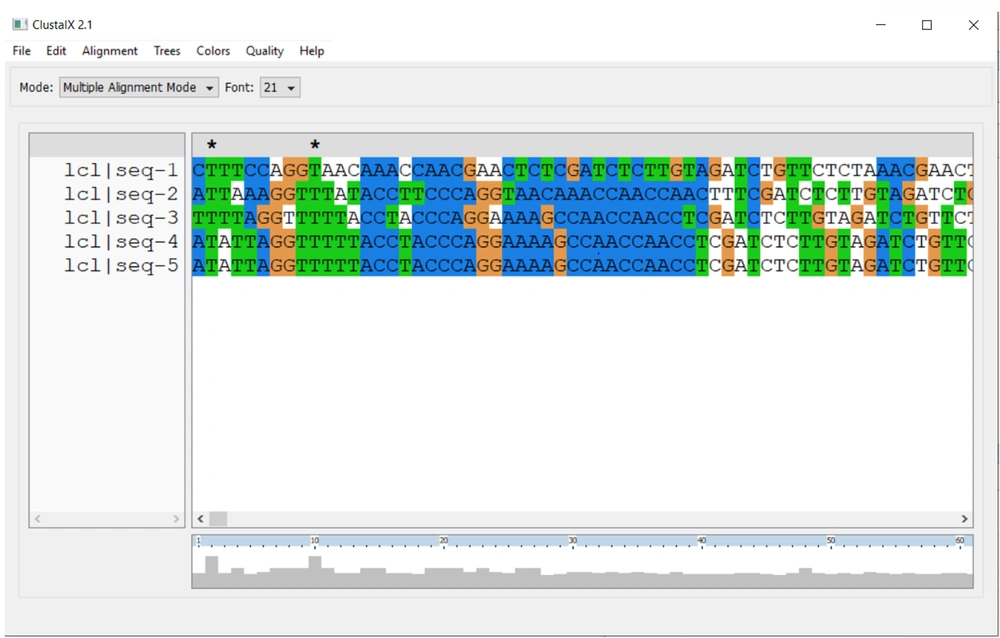
For aligning multiple sequences of betacoronaviruses, we used NCBI Genome Workbench (Multiple Sequence Alignment Viewer) and ClustalX (version 2.1). We can see the Receptor Binding Domain (RBD) and Polybasic Cleavage Site by scrolling across the sequence (25, 26) (Figure 8).
3.4.5. Phylogenetic Tree
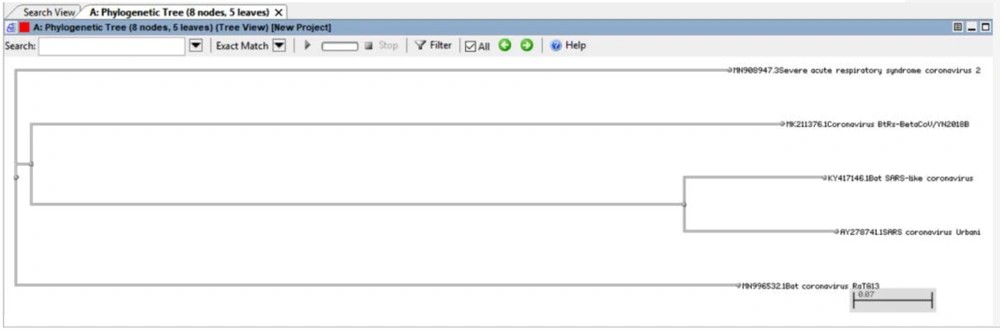
We can view the phylogenetic tree using the NCBI Tree Viewer (27) (Figure 9).
3.4.6. Viewing the Structure of Novel Coronavirus Spike Receptor-Binding Domain Complexed with Its Receptor ACE2
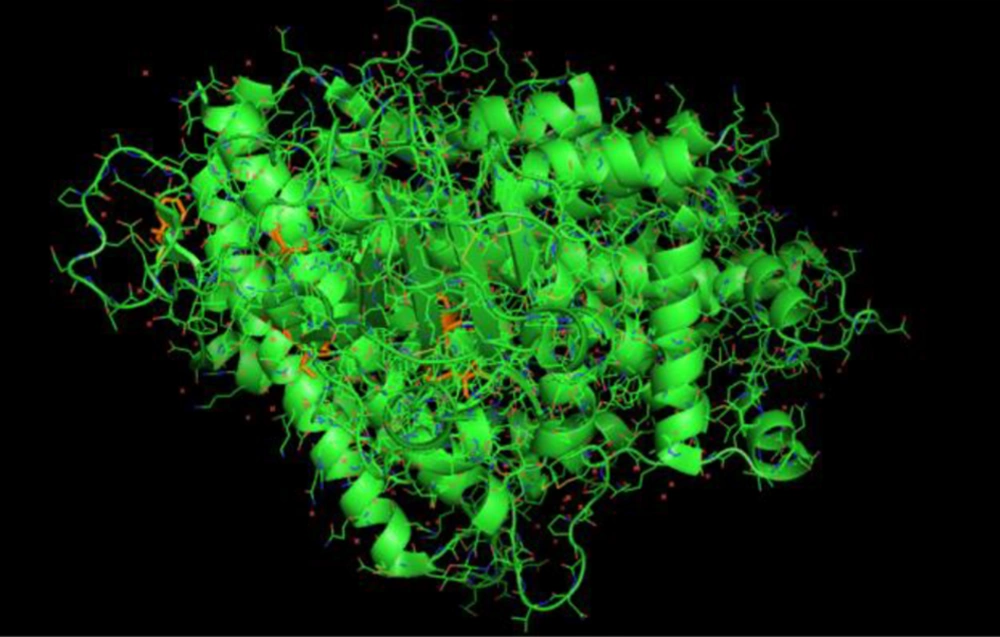
For this process, we obtained the SWISS MODELS from Protein Data Bank (PDB). The model chosen was the structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2. The structures were downloaded in PDB formats and processed in PyMol environment. By using this tool, we visualized and explored the molecular structure of SARS-CoV-2 spike glycoprotein (28-32) (Figure 10).
4. Discussion
4.1. Current Scenario in the Use of AI/ML Technologies in Vaccine Development for COVID-19
In the present circumstances, AI technologies are not widely employed by all researchers during vaccine and drug development. Only fewer machine learning models have been implemented for the spread of coronavirus. AI can forecast the rate in which an infection spreads in a particular region. This data can be further processed to help provide health officials the situation of a particular pandemic, thereby aiding the coronavirus response. However, there are many barriers involved such as availability of datasets, limited trained professionals, and resources. In AI and ML, every model is trained so that the solution to the problem is achieved. This is the basis of AI/ML-based researchers and scientists. We believe that this field could have an enormous impact on the current highly accelerating pandemic, COVID-19, that is taking over the world (33-37).
4.2. Applications of Artificial Intelligence and Accounts of Successful Trials
● Deep Learning-Based Drug Screening for Novel Coronavirus: This method was created by researchers in China using DenseNet to predict the interactions between proteins and ligands. This helps predict which drug combinations work preferably well in response to the virus (38-40).
● Predicting Antiviral Drugs that are Available Commercially to Control COVID-19: Here, a specific method called Molecule Transformer Drug Target (MT-DTI) is used (41).
● DeepMind and AlphaFold: DeepMind uses the AlphaFold library to predict the protein structures of COVID-19 (42, 43).
● Prediction of Critically Ill Patients in Wuhan Using ML Models: Artificial Intelligence scientists in Wuhan developed this method to identify the intensity of infections with factors such as age, gender, etc.
● Data based screening and Kalman filters for analysis of data.
● Further predictions of infection in a particular region (44-46).
4.3. Conclusions
Artificial Intelligence and Machine Learning is still subject to many criticisms, and many people are questioning its capacity to solve real-world problems. In other words, it is still an underutilized area in healthcare systems that needs to be developed to benefit millions of people. The main reason is that people prefer to handle every situation in a manual way. For instance, a hospital will prefer to handle its X-Ray or CT data in a manual way by doctors rather than train a model using CNNs to analyze the data. But if this technology is used to the maximum, it has the potential to create a revolution in the field of vaccine and drug development and benefit the whole of mankind.