1. Context
Reaching antibiotic resistance to critical status in recent years brings the World Health Organization (WHO) and the Centers for Disease Control (CDC) to declare that antibiotic resistance is becoming a pandemic (1). Therefore, alternative antimicrobial compounds are being developed to combat pathogens that already resist existing antibiotics (2). Pleuromutilins are a class of antimicrobial agents that inhibit protein synthesis. One of the pleuromutilin compounds approved by the Food and Drug Administration (FDA) in 2007 for human use is retapamulin (RET). It is a semi-synthetic derivative of pleuromutilin discovered by GlaxoSmithKline and known by the commercial name Altabox in the USA and Altargo in Europe (3-6). Topical RET ointment (1%) is used to treat impetigo caused by methicillin-susceptible Staphylococcus aureus (MSSA) and Streptococcus pyogenes.
Moreover, it has been shown that RET effectively treats secondarily infected wounds with negligible side effects. Furthermore, retapamulin shows fine in vitro activity against most facultative aerobic pathogens (2, 7). Besides, with rising resistance to mupirocin and fusidic acid in S. aureus and macrolides and clindamycin in S. pyogenes, retapamulin could act as a potential alternative drug for topical treatment and management of skin structure infections (2, 4, 8, 9). This paper is a brief review of the structure, action mode, metabolism, epidemiology of resistance, and the clinical use of retapamulin as a new antibiotic in the class of pleuromutilin anti-infective drugs.
2. Structure of Antibiotic
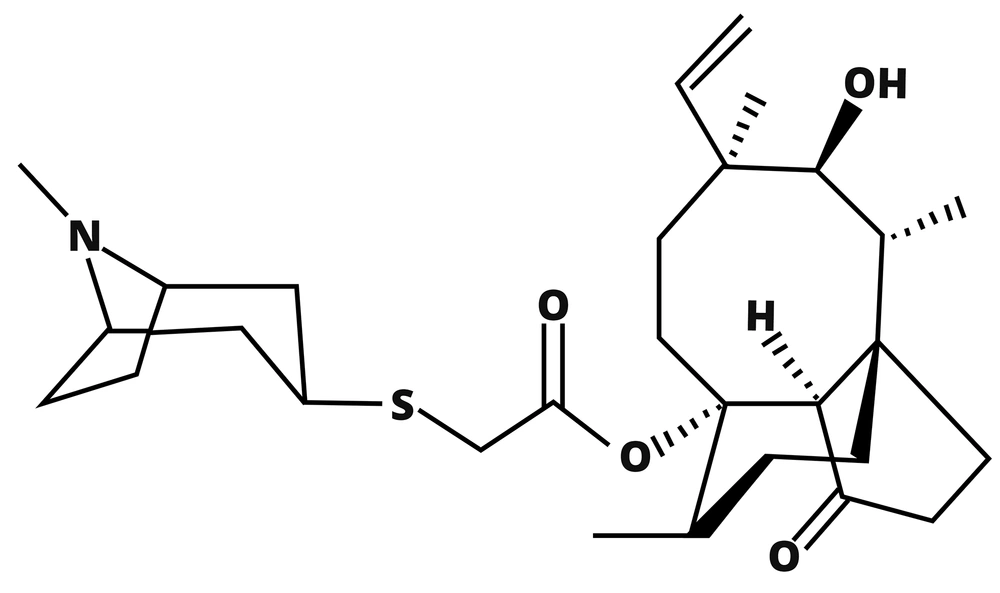
Pleuromutilin, first isolated from the fungi Pleurotus mutilis and P. passeckerianus in the 1950s by Kavanagh and his colleagues, is a tricyclic diterpenoid compound, as shown in Figure 1. It inhibits the synthesis of proteins by binding to the Peptidyl Transferase Center (PTC) of the ribosome (10). Pleuromutilin has a medium effect on Gram-positive bacteria and Mycoplasma, but the semi-synthetic derivatives of pleuromutilin with changes in C-14 have improved its activity against Gram-positive bacteria leading to new derivatives such as tiamulin, valnemulin, and retapamulin (RET). Only RET has been approved for human medicine, and the rest are used in veterinary for poultry and pigs. The C-14 side chain, in particular, can be considered the point of change in these compounds where the extensions can achieve extra hydrogen bonds between the drug and PTC and somehow overwhelm resistance mechanisms that are the result of mutations in the ribosome structure (5, 6, 11-14).
3. Mechanism of Action and Metabolism
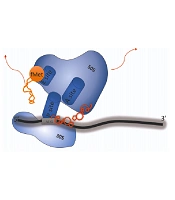
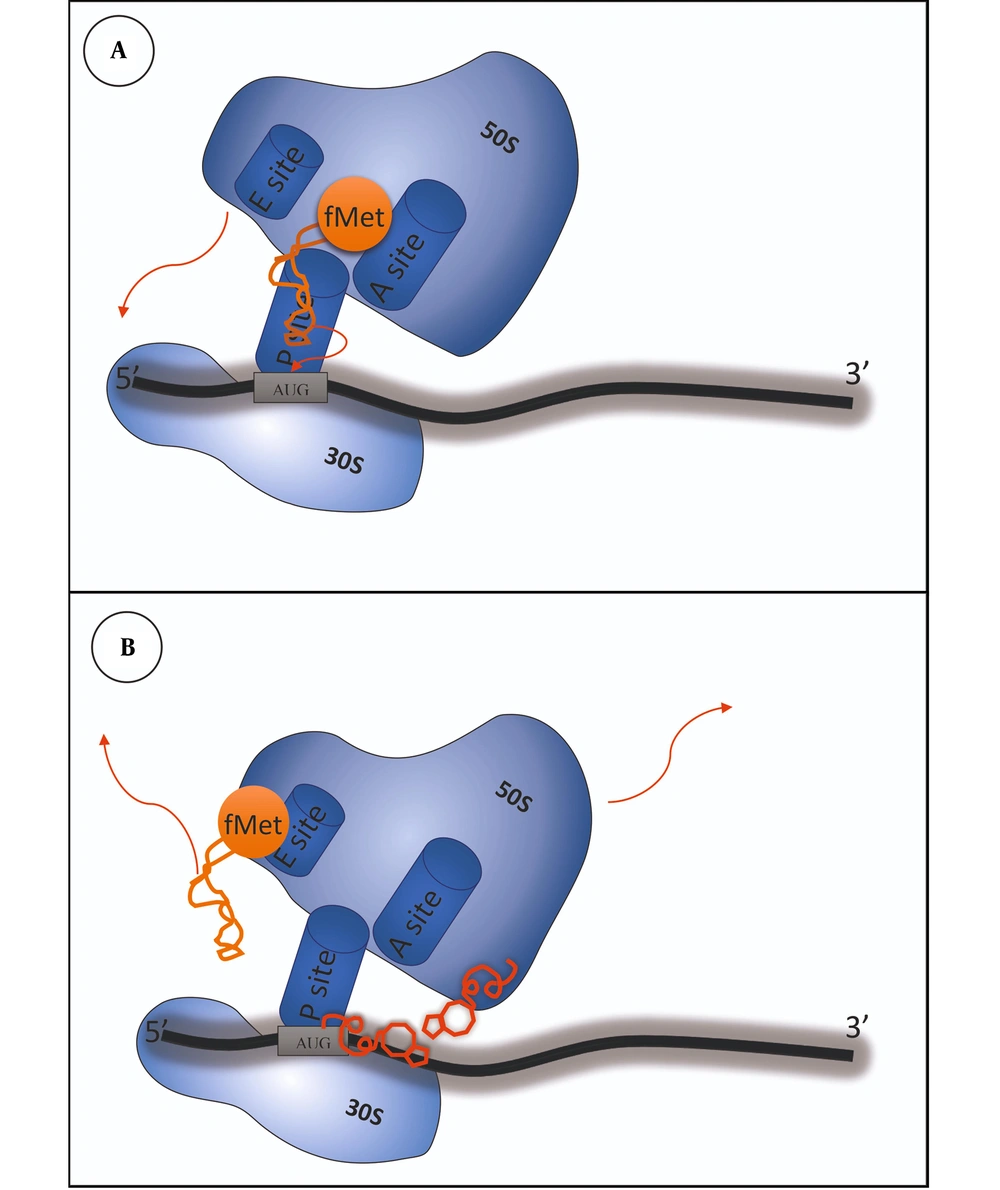
As previously mentioned, retapamulin plays its role by binding into the PTC of the ribosome, which causes the inhibition of active 50s ribosomal subunit and protein synthesis, contributing to growth inhibition and, in some cases, cell death (5). This mechanism differs from other antibiotic classes that inhibit protein synthesis by binding to the ribosome, like macrolides, lincosamides, phenicols, oxazolidinones, and streptogramins (8). As presented in Figure 2, retapamulin causes hydrogen-bonding interactions with the target. It also prevents protein synthesis by affecting three subjects: First by inhibiting the 50s subunit from binding to the 30s subunit, second by disrupting the peptidyl transferase and preventing fMet-tRNA from setting on the P site of the ribosome, and third by playing a role in impairing the tRNA binding to the P-site, leading to the relocation of tRNA on the ribosome so that tRNA would not be able to participate in the peptidyl transfer. This mechanism is unique and different from other antibiotics like macrolides and ketolides and diminishes the cross-resistance phenomenon (8). Furthermore, retapamulin has a high efficiency against isolates resistant to other inhibitors of protein synthesis and different classes of antibiotics like β-lactams, fusidic acid, erythromycin, mupirocin, azithromycin, levofloxacin, mupirocin, and quinolones. Additionally, RET has the advantage of not binding to eukaryotic ribosomes, thus not affecting mammalian protein synthesis (15). In summary, retapamulin, by making multiple interactions with the ribosome, plays its role in inhibiting bacterial growth (9, 16-18).
A, in a normal prokaryotic ribosome, translation begins with the interaction between the 30S subunit and the mRNA template when the initiation codon is placed at the P site. The tRNA charged with N-formylmethionine enters the p site. With the collaboration of initiation factors, the 50S subunit joins and conformational changes form the 70S subunit; B, when retapamulin exists in the cytoplasm, it can inhibit protein synthesis by causing hydrogen bonding interactions within the ribosomal complex, especially with PTC. Firstly, the conformational changes would not let the 50S subunit join the 30S and form the 70S subunit. Secondly, it prevents fMet-tRNA from setting on the P site of the ribosome, and then, it relocates the tRNA on the ribosome and would not let it take part in the peptidyl transfer.
With all good features, later on, it was found that retapamulin cannot be used for systemic infections because of the inadequate pharmacokinetic features such as low oral absorption, low water solubility, and rapid metabolism that results in a short lifetime (5). Data related to pharmacodynamics and pharmacokinetics of RET are very sparse, and the RET distribution in the human body is not well determined. In healthy adults who used 1% retapamulin ointment once daily, absorption, distribution, and metabolism were investigated. The results indicated that systemic exposure via the skin was shallow in topical use (5, 18). Furthermore, due to the topical use and low systemic exposure, the elimination from the body has not been analyzed yet, and the effect on normal flora is unknown. However, RET has been suggested to have too little interference with the gastrointestinal flora (5, 8). A study conducted by Sun et al. documented that cytochrome P450 enzymes metabolize RET in C-2 and C-8 positions, which accounts for low bioavailability and short efficacy of the drug (12). Due to its low exposure after topical use, interference with the metabolism of other P450 substrates seems unlikely (19).
4. In Vitro Susceptibility
Retapamulin was initially developed for topical treatment of skin infections caused by S. aureus and S. pyogenes. Also, it can be effective against many Gram-positive bacteria like Streptococcus pneumoniae, Coagulase-Negative Staphylococci (CoNS), Streptococcus agalactiae, Streptococci viridans, Propionibacterium acnes, Corynebacterium spp., and Micrococcus spp. (2, 20). However, it almost does not affect Gram-negative bacteria, except for Haemophilus influenza and Moraxella catarrhalis. It can be used against vancomycin-susceptible strains of Enterococcus faecium, while it does not affect E. faecalis (20). As the final point, RET shows good activity against anaerobic Gram-positive cocci and is as effective as Co-amoxiclav, imipenem, metronidazole, and clindamycin against Prevotella spp., Porphyromonas spp., and Fusobacterium spp. (2, 19). Notably, retapamulin can maintain its activity against other pathogenic strains that become resistant to methicillin, erythromycin, fusidic acid, mupirocin, azithromycin, and levofloxacin (20). Usually, RET is a bacteriostatic drug unless the concentration rises by 1000 times, becoming a bactericidal agent (19). The minimum inhibitory concentration (MIC) of retapamulin is shown in Table 1.
| Study and Species | MIC50 | MIC90 | MIC Range | Ref. |
|---|---|---|---|---|
| Pankuch et al. (2006) | (21) | |||
| Methicillin-susceptible Staphylococcus aureus | 0.06 | 0.125 | 0.03 - 0.125 | |
| Methicillin-resistant S. aureus | 0.06 | 0.125 | 0.06 - 0.25 | |
| Macrolide-susceptible Streptococcus pyogenes | 0.016 | 0.03 | 0.016 - 0.03 | |
| Macrolide-resistant S. pyogenes | 0.016 | 0.016 | 0.008 - 0.03 | |
| Harrington et al. (2016) | (4) | |||
| Methicillin-resistant S. aureus | 0.12 | 0.25 | 0.008 to ≥ 16 | |
| Jones et al. (2006) | (22) | |||
| Erythromycin-resistant S. aureus | 0.06 | 0.12 | ≤ 0.03 - 0.12 | |
| Mupirocin-resistant S. aureus | 0.06 | 0.12 | ≤ 0.03 - 0.12 | |
| Oxacillin-resistant S. aureus | 0.06 | 0.12 | ≤ 0.03 - 0.12 | |
| Erythromycin, mupirocin, and oxacillin susceptible S. aureus | 0.06 | 0.12 | ≤ 0.03 - 0.12 | |
| Mupirocin-resistant CoNS | ≤ 0.03 | 0.06 | ≤ 0.03 - 0.12 | |
| Oxacillin-resistant CoNS | ≤ 0.03 | 0.06 | ≤ 0.03 - 0.12 | |
| Mupirocin- and oxacillin-susceptible CoNS | ≤ 0.03 | 0.06 | ≤ 0.03 - 0.25 | |
| Erythromycin-resistant S. pyogenes | ≤ 0.03 | ≤ 0.03 | ≤ 0.03 - 0.06 | |
| Saravolatz et al. (2013) | (23) | |||
| Methicillin-resistant S. aureus | 0.12 | 0.12 | 0.06 - 0.12 | |
| Vancomycin intermediate S. aureus | 0.06 | 0.12 | 0.03 - 0.25 | |
| Vancomycin-resistant S. aureus | 0.06 | 0.12 | 0.03 - 0.25 | |
| Daptomycin-nonsusceptible S. aureus | 0.06 | 0.12 | 0.03 - 0.25 | |
| Mupirocin susceptible S. aureus | 0.06 | 0.12 | 0.03 - 16 | |
| Mupirocin low-level resistance S. aureus | 0.06 | 0.03 - 1 | ||
| Mupirocin high-level resistance S. aureus | 0.12 | 0.12 | 0.06 - 0.25 | |
| linezolid nonsusceptible S. aureus | 1 | 0.06 - 16 | ||
| Park et al.(2015) | (15) | |||
| Mupirocin-resistant S. aureus | 0.5 | 0.5 | 0.5 | |
| Rittenhouse et al. (2006) | (24) | |||
| S. epidermidis | 0.06 | 0.12 | 0.03 - 0.12 | |
| S. saprophyticus | 0.12 | 0.12 | 0.03 - 2 | |
| S. pyogenes | 0.008 | 0.016 | ≤ 0.004 - 0.12 | |
| S. agalactiae | 0.016 | 0.03 | 0.008 - 0.03 | |
| Viridans group streptococci | 0.03 | 0.12 | 0.008 - > 4 | |
| S. pneumonia | 0.06 | 0.12 | 0.03 - 0.12 | |
| Haemophilus influenza | 0.5 | 2 | 0.25 - 2 | |
| Moraxella catarrhalis | 0.3 | 0.3 | 0.016 - 0.06 | |
| Odou et al. (2007) | (2) | |||
| Bacteroides fragilis | 0.25 | 64 | 0.03 - 64 | |
| Bacteroides thetaiotaomicron | 16 | > 64 | ≤ 0.015 - > 64 | |
| Other Bacteroides spp. of the B. fragilis group | 2 | 32 | ≤ 0.015 - > 64 | |
| Clostridium perfringens | 0.125 | 1 | 0.03 - 1 | |
| Other Clostridium spp. | 0.5 | 64 | ≤ 0.015 - > 64 | |
| Propionibacterium acnes | 0.125 | 0.25 | ≤ 0.015 - 2 | |
| Goldstein et al. (2006) | (25) | |||
| P. acnes | ≤ 0.015 | 0.25 | ≤ 0.015 - 1 |
5. Mechanisms of Resistance to Retapamulin
Bacteria are capable of gaining resistance to antimicrobial drugs after exposure to sub-lethal doses. In a multistep resistance selection study, the authors passaged four MRSA strains in the presence of retapamulin with the initial MIC of 0.25 µg/mL. The final MIC at passage 55 was 8 µg/mL for three strains and 4 µg/mL for the leftover strains (26). This research indicated the potential for mutational resistance to RET, affecting the 50s ribosomal subunit. The resistance can be acquired by various mechanisms, including: (1) nucleotide substitution in rRNA or amino acid substitution in ribosomal proteins, thereby blocking peptide formation directly by interfering with substrate binding; (2) methylation of rRNA; (3) enzymatic inactivation of antibiotic; and (4) efflux pumps that expel antibiotics from the cell.
5.1. Protein Synthesis Inhibitor
The aforementioned antimicrobial agents bind to a well-conserved region of the ribosome, which is formed mainly of RNA. It is not expected that mutations occur in the conserved regions. Besides, many bacteria have multi-copy ribosomal RNA, which reduces the effect of a single mutation, and the copy number of 23s rRNA can be a limiting factor for drug-resistant mutants. Miller et al. investigated the mechanisms of linezolid and tiamulin cross-resistance in S. aureus and found that mutations in multi-copy ribosomal RNA (rrn) operons led to resistance to both classes of antibiotics (27). Also, the mutations in ribosomal proteins correlate with drug resistance. For example, the L3 S158 mutation (rplC) in S. aureus favors the resistance to tiamulin by deviating the conformation in the binding site (27-29).
5.2. Methylation of rRNA
The cfr gene is a multidrug resistance gene, often located on plasmids. It codes an enzyme that methylates the adenine residue at position 2503 in the enzymatic center of 23S rRNA, which is the binding site of antibiotics. The existence of this gene in bacterial cells can cause multi-resistance to phenicols, lincosamides, macrolides, oxazolidinones, pleuromutilins, and streptogramin A (known as PhLOPS phenotype) (30). Several studies have described the presence of cfr in unexpected outbreaks of linezolid-resistant Staphylococci and considered it an emerging global health threat, which can be associated with pleuromutilins resistance (31, 32).
5.3. Enzymatic Inactivation of Antibiotic
Until now, there was not any report of an antibiotic-inactivating enzyme that especially makes the pleuromutilins inoperative. Such enzymes can modify the structures of antibiotics by hydrolyzing, transferring diverse groups, and performing oxidoreductase activity that hurdles the target affinity, decompose the antibiotic, or block the cellular uptake (33). Covalent modification of antibiotics can perturb target affinity, block cellular uptake, trigger efflux mechanisms, or lead to the decomposition of the antibiotic.
5.4. Efflux Pumps
Efflux pumps are active transporters that cause multiple-drug resistance in pathogenic bacteria. They can increase the MICs of their substrates, but not necessarily to the levels indicative of clinical resistance. Efflux can be the fastest and most effective resistance mechanism for antibiotics that act inside the bacterial cell. As known, Vga (A), Vga (C), and Vga (E) are members of the ATP-binding cassette (ABC) family of proteins that export streptogramins, lincosamides, and pleuromutilins (34-38). Harrington et al. analyzed a single S. aureus isolate with an elevated RET MIC by polymerase chain reaction (PCR) for the vgaA gene. The sequenced amplified product was 100% identical to vgaA of S. aureus, and they suggested that the elevated MIC might be linked to the acquisition of vgaA (4). In a study on S. agalactiae isolates from the USA, Hawkins et al. found out that lnu (B) is invariably present in combination with lsa (E) on the same transposable element (TE) and transforms cross-resistance to lincosamides, streptogramins A, and pleuromutilins among the isolates (39). Besides, lnu encodes the lincosamide O-nucleotidyltransferase, which enzymatically inactivates the antibiotic (40). Also, lsa, which codes for an ABC transporter, was first discovered in E. faecalis (41). Since this TE has been discovered in S. aureus (42), these genes are expected to act synergistically and produce cross-resistance in S. aureus (43, 44). Recently, Schuster et al. evaluated the impact of RND-type transporters on the susceptibility to available oxazolidinones and pleuromutilins in E. coli. They showed different pathways for oxazolidinones and pleuromutilin derivatives through resistance-nodulation-division (RND)-type transporters, depending on the molecular weight of the components (45). Unexpectedly, not all combined mechanisms increase the resistance to ribosome-targeting antibiotics. For example, the degree of resistance achieved from Cfr methyltransferase depended on L3 mutations, and some of them could decrease the effect of methylation and lower the MIC (46).
6. Epidemiology of Retapamulin Resistance
The data on the worldwide prevalence of RET resistance between clinical isolates of Gram-positive cocci are scanty, with rates of resistance ranging from less than 1 to 2.6% (brief data are shown in Table 2). The point is that RET acts effectively on MRSA in vitro but shows poor outcomes in clinical trials. Most studies reported negligible RET-resistance rates among MRSA isolates, which may resist the other antibiotics such as vancomycin, linezolid, mupirocin, and fusidic acid. Epidemiologic findings suggest that RET-resistant isolates cross-resist with linezolid frequently (23, 47, 48). Furthermore, by examining the presence of the smr, vga, and cfr genes by PCR and sequencing the 23S rRNA and the gene encoding ribosomal protein L3, rplC, in resistant isolates, the authors investigated whether a particular gene or mutation is responsible for the RET-resistance phenotype.
| Year | Region | Population | Susceptibility | Ref. |
|---|---|---|---|---|
| 2008 | UK | Strains were submitted to the HPA Centre from the UK | 99.9% (664/663) | (6) |
| 2008 | USA | 390 streptococcal strains and 344 staphylococcal strains collected from recent clinical isolates | 100% (390/390); 95.6% (344/329) | (49) |
| 2011 | Spain | Collection of clinical isolates that 18 of them were linezolid-resistant and 10 were MRSA. | 52.6% (38/20) | (48) |
| 2013 | USA | A diverse group of strains, including mupirocin, VRSA, VISA, and daptomycin-non-susceptible isolates | 97.4% (155/151) | (23) |
| 2014 | USA | Pediatric patients with a median age of 27 months | 86.0% (400/344) | (50) |
| 2015 | Korea | Clinical MRSA isolates from patients of tertiary hospitals | 100% (497/497) | (15) |
| 2018 | Singapore | Non-duplicate MRSA clinical isolates | 100% (200/200) | (51) |
Over time, the expanding use of topical ointments containing retapamulin caused a slight rise in the prevalence of resistance to RET in the USA, where this alternative is used in treatment regimens. However, in Asia, S. aureus clinical isolates are pretty susceptible. In addition, data have shown that the prevalence of mupirocin-resistant isolates has dramatically increased via gene transfer between strains, and thus, retapamulin as a promising alternative can be applied to eradicate these resistant isolates (Table 2).
7. Clinical Treatment
By increasing resistance to well-known antibiotic classes such as beta-lactams, the interest in pleuromutilins has revived. In 2007, retapamulin was the first pleuromutilin that got approval for clinical use in humans in the form of ointment 1% (Altabax) (52). For the treatment of localized impetigo, a cutaneous infection due to S. aureus and S. pyogenes, topical antibiotics with fewer side effects are considered the drug of choice. Two clinical trial studies (7, 9) have proven the efficacy and safety of retapamulin ointment against bacteria, showing no statistical differences between retapamulin and fusidic acid for impetigo treatment (53). Although the use of retapamulin for treating infections with other etiologies is not recommended, evidence indicates the excellent potential of this antibiotic for the treatment of skin and soft-tissue infections related to anaerobe bacteria (2).
Furthermore, other clinical trial studies (54, 55) revealed that RET is an effective drug for treating secondarily infected traumatic lesions (SITLs), which has been non-inferior to oral cephalexin. In a recent post-marketing surveillance (PMS) study, the effectiveness rate was 96.1% (1,697 out of 1,765 subjects) and the incidence of serious adverse events was 0.28% (56), while adverse events were primarily local, including irritation, burning, pruritus, pain, erythema, eczema, and contact dermatitis. Despite these advantages, the clinical efficacy of topical retapamulin for the treatment of MRSA infections is doubtful compared to other antibiotics like linezolid and cephalexin (57, 58). Nevertheless, no studies have yet indicated the exact cause of efficacy reduction in MRSA strains.
Nasal S. aureus (including MRSA) carriage is one of the most important causes of the dissemination of nosocomial infections in health care settings. Given the high rate of nasal MRSA carriage (59), eradicating MRSA nasal carriage from patients and hospital staff can help control outbreaks. Mupirocin has been applied for the nasal decolonization of the carriers since the 1980s (60). The mupirocin-resistant phenotype emerges when the mutation of chromosomal gene ileS-2 (mupA) occurs, or the bacteria gain the alternative gene by transferable plasmids (61). Therefore, new therapies are needed to replace mupirocin (62). Harrington et al. evaluated the activity of RET against MRSA isolates by in vitro experiments and noticed that 99% of them were susceptible (4). A phase-IV clinical trial of retapamulin as a topical decolonizing agent for mupirocin-resistant MRSA was conducted in 2011 (NCT01461668), reporting the efficacy of retapamulin in eight out of 25 (32%) participants versus four out of 25 (16%) in the placebo group. After a three-year follow-up, no adverse effect was reported from subjects. The authors also reported that RET significantly reduced mupirocin-resistant MRSA strains for only one week, introducing it as an excellent alternative to mupirocin in temporary risk periods such as surgeries and ICU stays (63).
8. Other Pleuromutilin Derivatives for Clinical Use
Among many pleuromutilin semi-synthetic analogs, BC-3781, best known as lefamulin, is the first pleuromutilin evaluated for clinical systemic uses. Lefamulin is developed by Nabriva Therapeutics for oral and intravenous administration. It has been currently passed phase III clinical trials for the treatment of community-acquired bacterial pneumonia (CABP), acute bacterial skin and skin structure infections (ABSSSIs), and sexually transmitted infections (64-66). Lefamulin has potent activity against Gram-positive, Gram-negative, and atypical pathogens associated with respiratory tract infections (65, 67). Previous studies have shown that lefamulin can reach a high concentration in the cell and has rapid accumulation into murine macrophages while its potency would not be affected by lung surfactants (65, 68). Also, it shows rapid plasma absorption and fast, predictable penetration into soft tissue and lung epithelial lining fluid (ELF) (66, 69).
Furthermore, it indicates that lefamulin is potentially as effective as vancomycin in treating ABSSSIs caused by Gram-positive pathogens (66, 70). Current data demonstrate that lefamulin is safe and well-tolerated, and since it reaches the therapeutic concentrations in the ELF, it can be used to treat adults with CABP (66). Additionally, like other pleuromutilins, it inhibits protein synthesis with a unique mechanism, so the chance of cross-resistance with other antibiotics would be minimized. The FDA approved lefamulin in August 2019 based on results from the Lefamulin Evaluation Against Pneumonia (LEAP 1) (NCT02559310) and LEAP 2 (NCT02813694) clinical trial studies, which compared five-day oral lefamulin with seven-day oral moxifloxacin for the treatment of CABP. The efficacy of lefamulin was non-inferior to that of moxifloxacin in adults with CABP and was generally safe and well-tolerated (71, 72). Lefamulin empiric monotherapy gives high response rates to typical and atypical CABP pathogens, and consequently, clinicians can use it as a potential alternative for the treatment of CAPB when antibiotic resistance limits the primary antibiotics for treatment.
9. Conclusions
Increased antibiotic resistance has been one of the severe issues in recent studies, prompting research to discover and introduce new antimicrobials with greater efficacy and accuracy against resistant superbugs. The components of pleuromutilins have been used in veterinary medicine for many years, and recently a semi-synthetic derivative called retapamulin has introduced itself as an alternative topical antibiotic. Experiments have shown that it was as effective as common topical agents and superseded them due to the increased rate of "resistance" among older antibiotics, and this point remains one of the most common indications for using novel antibiotics. Although it is not helpful for systemic use owing to rapid hepatic metabolism, it can be used in topical ulcers with Gram-positive etiology. It also can temporarily stop MRSA colonization in patients and hospital staff. Research on bacterial protein synthesis and ribosome profiling can use RET due to its bacteriostatic properties (73, 74), and since it has a different binding site than have routine antibiotics such as chloramphenicol, these studies may give us new data in this field. Finally, a more recently pleuromutilin derivative with systemic usability, lefamulin, is undergoing clinical trials. In this review, we attempted to examine the derivatives usable in humans briefly and explore their structures, action mode, metabolism, possible ways of resistance, resistance rates, and clinical use to explain and highlight the valuable points of these antibiotics. It is suggested that future in vivo studies and clinical trials focus on protecting the RET structure against oxidation and examine its potential for eradicating other etiologies of wound infection.