1. Background
COVID-19 is an acute respiratory infectious disease caused by the virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1). In January 2020, the World Health Organization (WHO) declared the COVID-19 outbreak a pandemic. The clinical outcomes of COVID-19 range from mild symptoms to severe complications and ultimately death, making it a significant global health concern (2, 3). The rapid spread of the disease has resulted in shortages of medical equipment and burnout among healthcare workers (4, 5). As of August 16, 2023, there have been 770,437,327 confirmed cases of COVID-19 globally, including 6,956,900 deaths (6). In the United States, from January 2020 until June 2023, there were 103,436,829 confirmed cases of COVID-19, with 1,127,152 deaths (6).
The main symptoms of COVID-19 include fever, cough, and shortness of breath, and the virus has high transmission and prevalence rates. Other symptoms may include tiredness, loss of taste or smell, muscle aches, chills, sore throat, runny nose, headache, chest pain, pink eye, nausea, vomiting, diarrhea, and rash (7). The severity of COVID-19 symptoms can range from very mild to severe. Some individuals may have no symptoms at all but can still spread the virus through asymptomatic transmission. The virus spreads via respiratory droplets released when someone coughs, sneezes, breathes, sings, or talks. COVID-19 is highly contagious and has led to a rapid pandemic that poses a serious threat to global public health (8, 9).
Due to the limited availability of diagnostic tests, accurately diagnosing COVID-19 remains one of the major challenges in managing this disease (10, 11). Alongside polymerase chain reaction (PCR) diagnostic tests, chest computed tomography (CT) scans are a significant diagnostic method for detecting the virus and monitoring the progression of the disease. Although chest CT scans may yield “false positives” in some cases, they remain a powerful tool for disease diagnosis. According to specialist reports, three types of abnormalities on CT scan images indicate COVID-19 infection: (1) Ground glass opacification, (2) consolidation, and (3) pleural effusion (12). Developing new tools for the improved detection of these irregularities in radiology images can greatly aid in controlling and managing COVID-19 (11).
Recently, the application of artificial intelligence and machine learning methods has been recognized as an efficient approach in the medical field. For example, Causey et al. reported an algorithm for predicting lung cancer using CT scan images and deep learning approaches, achieving an accuracy of 78% (13). Ardakani et al. developed eight machine learning models to distinguish COVID-19 from other non-COVID-19 lung diseases, achieving a ROC AUC of 0.994 for COVID-19 detection using their recent model (14).
2. Objectives
Given the importance of timely disease diagnosis, this study employed machine learning methods to predict the mortality of patients with COVID-19 infection.
3. Methods
3.1. Type of Study, Study Design, and Patient Selection
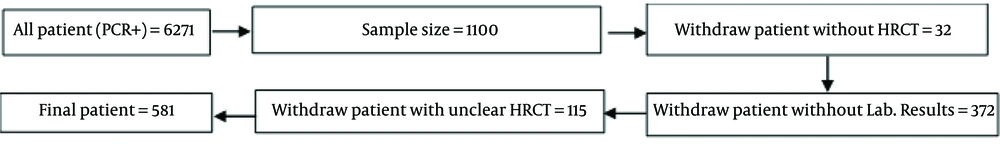
This cross-sectional study was conducted on 1,100 confirmed COVID-19 patients who were hospitalized at Imam Reza or Qaem hospitals under Mashhad University of Medical Sciences between December 2019 and December 2021. The 1,100 patients were selected using systematic random sampling. If a patient’s high-resolution computed tomography (HRCT) information was unavailable, another patient was selected as a replacement. Demographic information, chronic disease history, imaging findings, vital signs, and laboratory results were collected from the patients’ electronic medical records at the time of admission. The flowchart detailing patient selection and data collection is presented in Figure 1.
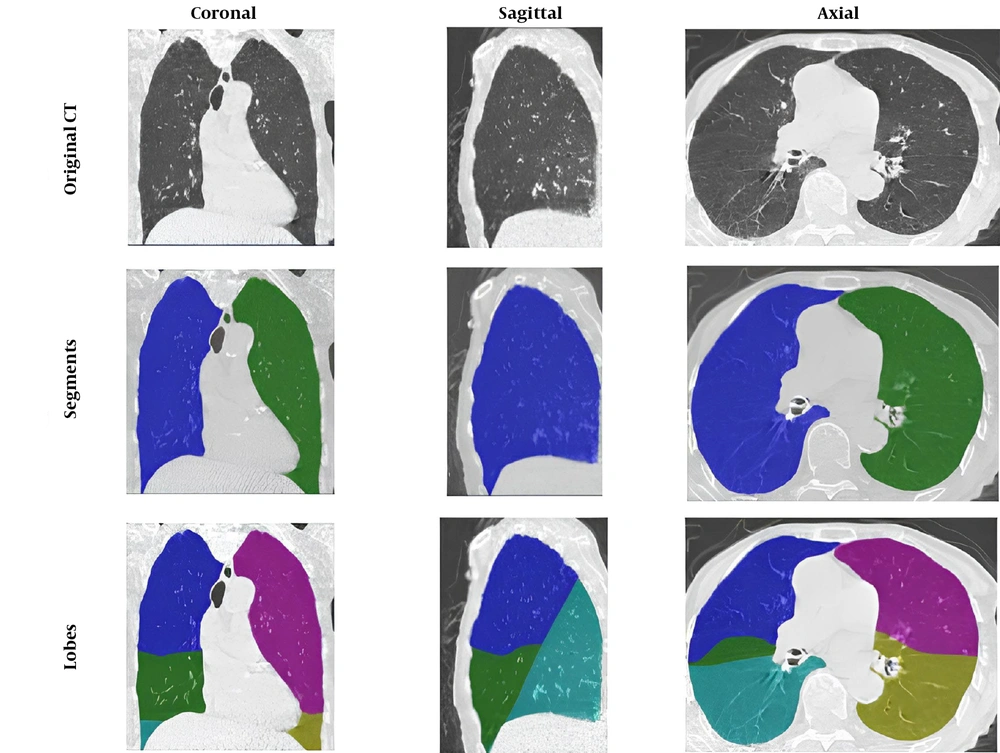
The patients' tissue and lung size values from CT scan images were analyzed using the Pulmonary Toolkit package in MATLAB software (Figure 2). Among the remaining 953 samples, 604 patients recovered, while 349 patients died. After incorporating laboratory information, 581 patients with complete data were included in the study.
3.2. Inclusion and Exclusion Criteria
The inclusion criteria for this study were as follows: Patients who were hospitalized with a positive PCR test and had HRCT images available. Patients without HRCT images were excluded from the study.
3.3. Statistics and Machine Learning Algorithm
Numerical variables were summarized using mean and standard deviation. To enhance the performance of data mining models and to determine the relationships between variables affecting COVID-19 mortality, t-tests, Mann-Whitney U tests, and chi-squared tests were employed. These tests were used to identify significant associations between variables and patient outcomes (death or recovery). A P-value < 0.05 was considered statistically significant. Variables with a significant relationship to the response variable were identified as risk factors.
In the machine learning model, the chi-squared (χ²) feature selection algorithm was used to identify significant variables, accommodating both quantitative and qualitative variables. This algorithm is based on the χ² statistic. The χ² value for r, defined as the difference in k classes, is represented as follows (15).
nij: Is the feature of jth case
ni*: The number of ith feature at all features
n*j: The number of samples in jth class
n = Sample size
In this study, Adaptive Boosting (AdaBoost) was used as a machine learning method to predict COVID-19-related conditions, considering the type and quality of the data (Table 1). AdaBoost is based on decision tree algorithms and works by combining a high-accuracy predictor with variables that have relatively weaker accuracy (16, 17).
| Variable | Predicted Values | |
|---|---|---|
| Actual values | Death (+) | Recover (-) |
| Death (+) | TP | FN |
| Recover (-) | FP | TN |
To evaluate the model's performance, 10-fold cross-validation was implemented. This statistical method for machine learning divides the dataset into training and validation sets across multiple iterations, ensuring that each data point is tested. Performance metrics, including accuracy, precision, recall, F-score, ROC AUC, and MCC, were calculated to assess the effectiveness of the predictive models (Table 2).
| Performance Metrics | Formulas |
|---|---|
| Accuracy | |
| Precision | |
| Recall | |
| F-score | |
| Matthew’s correlation coefficient |
In summary, the eligibility criteria and statistical methods were as follows:
3.4. Eligibility Criteria
The eligibility criteria for inclusion in the study were:
- Patients must have been hospitalized with a confirmed diagnosis of COVID-19 via PCR.
- Availability of HRCT images was required; patients without these images were excluded.
- A total of 32 patients were excluded due to missing HRCT images. From the remaining 1,068 DICOM images, 115 samples were discarded due to unclear imaging. Ultimately, 581 patients with complete data were included in the analysis.
3.5. Statistical Methods
To analyze the data and determine relationships between variables affecting COVID-19 mortality, several statistical methods were employed:
- t-tests, Mann-Whitney tests, and chi-squared tests were used to identify significant relationships between variables and patient outcomes (death or recovery), with a significance level set at P < 0.05.
- Variables that demonstrated a significant relationship with mortality were classified as risk factors for prediction purposes.
- Adaptive Boosting (AdaBoost), based on decision tree algorithms, was utilized to enhance prediction accuracy by combining strong predictors with weaker ones.
- To evaluate the model's performance, 10-fold cross-validation was implemented. This method divides the dataset into training and validation sets across multiple iterations, ensuring that each data point is tested.
- Performance metrics, including accuracy, precision, recall, F-score, ROC AUC, and MCC, were calculated to assess the effectiveness of the predictive models.
3.6. Mitigation Strategies for Bias
Mitigation strategies for various types of biases in this study included:
- Using data from referral hospitals and employing systematic sampling methods to reduce selection bias.
- Involving expert clinicians and methodologists to minimize measurement biases.
The analysis may not fully account for confounding factors that could influence patient outcomes, such as variations in treatment protocols or differences in healthcare access among different populations. A total of 32 patients were excluded due to missing HRCT images, and an additional 115 samples were discarded due to unclear imaging. These exclusions could result in the loss of potentially relevant data and may impact the generalizability of the findings.
While the study provides valuable insights into COVID-19 patient outcomes, caution should be exercised when applying its findings to broader populations due to differences in demographics, healthcare practices, and the evolving treatment landscape.
4. Results
The study was conducted on 581 patients, of whom 295 were male with an average age of 50.3 years, and 286 were female with an average age of 50 years. Of these, 199 patients were in the mortality group, and 382 were in the recovery group. The descriptive statistics are presented in Table 3.
| Variables | Death (n = 199) | Non-death (n = 382) | P-Value |
|---|---|---|---|
| Age | 66.57 | 57.34 | < 0.001 |
| Gender | 0.31 | ||
| Male | 113 (56.7) | 182 (47.6) | |
| Female | 86 (43.2) | 200 (52.3) | |
| Comorbidities | |||
| Nausea | 8 (4) | 24 (6) | 0.276 |
| Cancer | 8 (4) | 2 (0.5) | 0.002 |
| Diabetes | 27 (13) | 60 (15) | 0.493 |
| Asthma | 3 (1.5) | 4 (1) | 0.629 |
| Heart disease | 10 (5) | 30 (7) | 0.201 |
| Chronic kidney disease | 4 (2) | 5 (1) | 0.516 |
| Chronic lung disease | 5 (2) | 5 (1) | 0.29 |
| Hypertension | 38 (19) | 79 (20) | 0.651 |
| PO2 | 83.7 | 87.7 | < 0.001 |
| Image processing result | |||
| Percent of air | 61.18 | 65.22 | < 0.001 |
| Volume of air, cm3 | 1490.27 | 1698.42 | 0.005 |
| Percent of emphysema | 10.97 | 11.58 | 0.403 |
| Mean density, HU | -611.81 | -652.22 | < 0.001 |
| Percent of tissue | 38.81 | 34.77 | < 0.001 |
| Emphysema | 896.49 | 911.89 | 0.027 |
| Laboratory finding on admission | |||
| White blood cell, × 1000/mL | 8.41 | 7.24 | 0.236 |
| Red blood cell, × 1000/mL | 9.81 | 6.26 | 0.003 |
| LDL | 104.2 | 68.75 | 0.338 |
| Ferritin, ng/mL | 689.08 | 541.74 | 0.027 |
| FBS | 156.10 | 209.57 | 0.069 |
| D-dimer, ng/mL | 325.8 | 167.2 | < 0.001 |
| C-reactive protein, mg/dL | 10.8 | 6.6 | < 0.001 |
| Percent of Lymphocytes | 14.15 | 9.65 | < 0.001 |
a Values are expressed as No. (%) unless otherwise indicated.
The average age of deceased patients was 66.57 years, compared to 57.34 years for recovered patients. In the comorbidities subgroup analysis, only patients with cancer showed a significant difference (P-value = 0.002, Table 3). Additionally, the comparison of the mean SpO₂ Index between deceased and recovered patients was statistically significant (P-value = 0.002).
From the image processing results, the mean lung density, the percentage of air in the lungs, and the volume of air in the lungs were statistically different between the deceased and recovered groups (P-value < 0.001), P-value < 0.001, and P-value = 0.005, respectively). Furthermore, the emphysema Index in the recovery group was significantly higher than in the deceased group (P-value = 0.002).
In laboratory findings, there were significant differences in red blood cell (RBC) counts, lymphocyte levels, C-reactive protein (CRP), ferritin, and D-dimer levels between deceased and recovered patients.
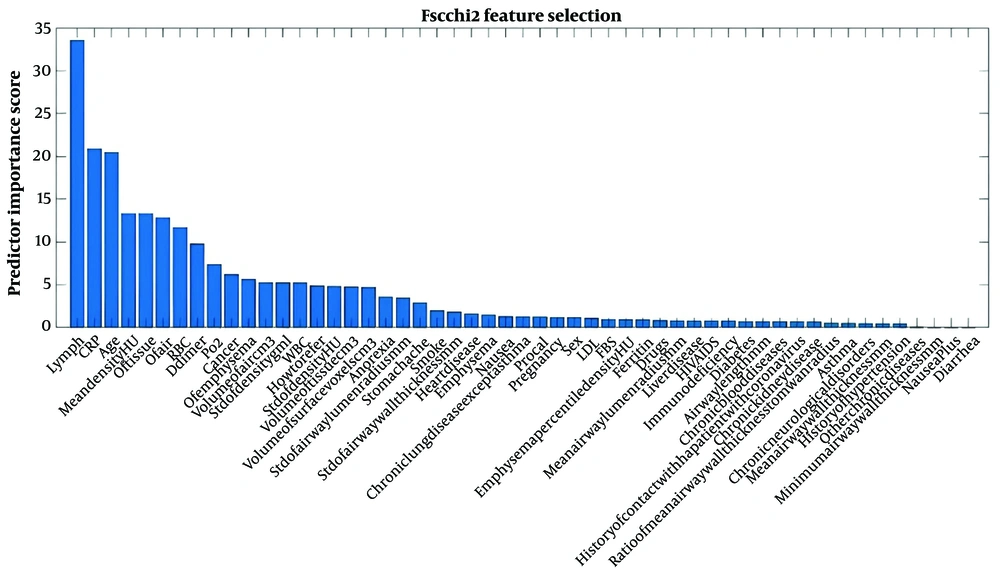
The Adaptive Boosting (AdaBoost) model was fitted to the data to predict treatment outcomes, and the three variables with the most significant impact on prediction are shown in Figure 3. In the final model, 10 variables—including lymphocytes, CRP, age group, mean tissue density, RBC, D-dimer, pO₂, cancer, and emphysema—were identified as having the most significant impact on prediction and were included in the analysis.
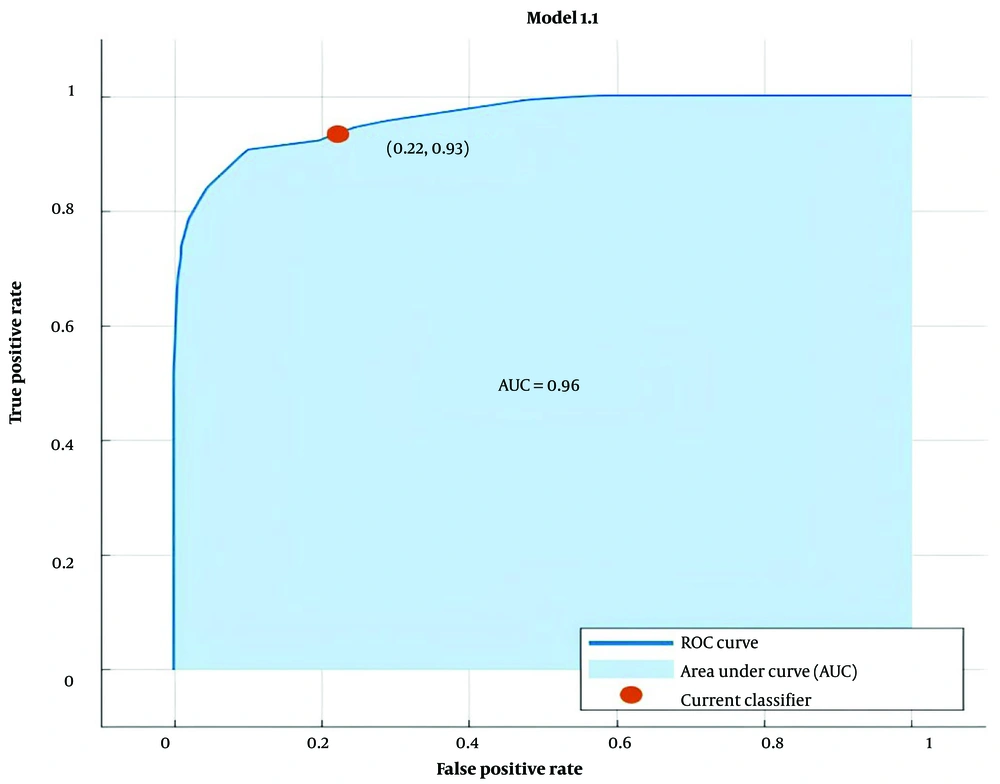
After fitting the model to predict treatment outcomes, the ROC curve was plotted to evaluate the model, yielding an AUC of 0.96 (Figure 4). Table 4 presents the confusion matrix for the AdaBoost model in predicting the outcomes of hospital care for COVID-19 inpatients. The evaluation metrics of the model are displayed in Table 5. The accuracy and precision of the model were 0.88 and 0.89, respectively. Predictive models like this one aim to maximize the agreement between predicted and actual values regarding recovery and mortality. Matthew's correlation coefficient (MCC) showed a value of 0.73 (Table 5).
| Variables | Predicted Survival | Predicted Death |
|---|---|---|
| Actual survival | 357 | 25 |
| Actual death | 44 | 155 |
| Index | AdaBoost |
|---|---|
| Accuracy | 0.88 |
| Precision | 0.89 |
| F-measure | 0.84 |
| Recall | 93.1 |
| Matthew’s correlation coefficient | 0.73 |
5. Discussion
This study presents a retrospective analysis of patient data to predict the mortality of COVID-19 patients hospitalized in referral hospitals between 2010 and 2021. Machine learning algorithms were applied to predict disease outcomes based on clinical data from hospitalized patients.
Lai et al. used the Adaptive Boosting algorithm to identify the most effective variables for predicting mortality in COVID-19 patients. Their findings revealed that lymphocyte counts were significantly lower in patients with severe COVID-19 compared to those with mild cases (18).
Lymphocyte count and CRP are two important variables in predicting the risk of death in patients with COVID-19. Several studies have shown that lymphocyte count serves as a universal predictor of health outcomes in COVID-19 patients (19).
Windradi et al. have indicated that CRP, as an acute-phase protein, is an effective marker for predicting severe COVID-19 (20). In a meta-analysis study, it was demonstrated that CRP is a significant variable in distinguishing between severe and mild cases of COVID-19 (21).
In the present study, we found that RBC was an effective variable for predicting the risk of death in COVID-19 patients. Hemoglobin in RBCs is considered an important biomarker, reflecting oxygen levels in the blood and serving as a significant variable in predicting COVID-19 mortality (22). Thomas et al. showed that RBC counts were significantly higher in COVID-19 patients compared to healthy individuals (23).
Additionally, age has been identified as a crucial variable for predicting COVID-19 mortality (24, 25). Bonanad et al. conducted a meta-analysis of 611,583 COVID-19 patients across five continents to investigate mortality rates among different age groups. They found that the mortality rate for individuals under 50 years old was 1.1%, and this rate increased with age, peaking in individuals aged 80 years or older (26). Another study found that individuals aged 55-64 years had an 8.1-fold higher COVID-19 mortality rate than those under 55 years of age (27). These findings suggest that age is a significant predictor of COVID-19 mortality. As age increases, the mortality rate also rises, with the highest mortality rates observed in patients aged 80 years and above (24).
Lyu et al. aimed to evaluate the severity of COVID-19 based on HRCT images. They found that the mean lung density, measured on the HU Scale, was higher in patients with severe COVID-19 compared to healthy individuals (28). In our study, the mean lung density in deceased individuals was also found to be higher than in those who recovered. Notably, the diagnostic value of CT scanning in assessing lung density has already been well-established and is considered preferable to other subjective visual examinations (29).
The data suggest that lung density is a potential imaging tool for assessing the severity of COVID-19, and its results can be valuable for identifying patients at risk of severe disease progression (30). However, further studies are necessary to validate the clinical utility of lung density analysis in managing COVID-19.
Additionally, we observed that the average D-dimer level was significantly lower in recovered individuals compared to deceased patients (P-value = 0.001). D-dimer is a blood biomarker that plays a critical role in predicting outcomes for patients with COVID-19 (31). One study indicated that the mean D-dimer level in patients with mild COVID-19 was approximately one-sixth of that in patients with severe disease (32).
It has also been demonstrated that patients with malignancies are at a higher risk of COVID-19 infection and severe complications due to their immunocompromised state (33). Similarly, other studies have reported an increased rate of COVID-19-associated mortality among cancer patients (34, 35).
The risk of severe COVID-19 outcomes increases with age, and patients with malignant tumors are at a higher risk for severe illness due to their underlying medical conditions (36). During the COVID-19 pandemic, cancer patients have had limited access to medical facilities and services, which has increased the likelihood and severity of their conditions (37). In our study, a significant difference was observed in the proportion of cancer patients between the deceased and recovered groups (P-value = 0.02). Patients with malignancies are at higher risk for severe complications and mortality from COVID-19 due to their immunocompromised state and underlying medical conditions. Vaccination has been shown to help reduce deaths and severe illness from COVID-19, as well as to decrease transmission in these patients (38).
In recent studies, predicting the severity and mortality of COVID-19 has been a major focus. Several studies have explored the relationship between COVID-19 and mortality, including excess mortality due to COVID-19, as well as machine learning models to predict mortality and critical events in COVID-19 patients. In a study by Akhtar et al., 10 machine learning algorithms were used to predict COVID-19 infection based on CBC results (39). According to their results, the highest accuracy (100%) in predicting infection was achieved by three algorithms: Random Forest, K Nearest Neighbor (KNN), and kStar. These findings suggest that machine learning algorithms can be useful in predicting COVID-19 infection based on CBC results. Further research is needed to establish the clinical utility of these algorithms in managing COVID-19. Moulaei et al. conducted a study on 1500 COVID-19 patients to predict mortality using various machine learning models. Their results showed that the ML and RF methods had the highest accuracy (> 80%) (1). In another study, Zakariaee et al. assessed the performance of four machine learning algorithms (LR, RF, SVM, and XGBoost) and found that XGBoost had the best performance in terms of AUC (40).
Schiaffino et al. conducted a study on 897 hospitalized COVID-19 patients to predict in-hospital mortality using HRCT scans. The algorithms used in this study were Support Vector Machine (SVM) and multi-layer perceptron (MLP). The area under the ROC curve for the SVM and MLP models was 0.74 and 0.84, respectively (41). Nuthalapati et al. used deep learning methods to predict mortality or hospitalization in the intensive care unit (ICU) for COVID-19 patients. Other variables, such as HRCT images and electronic health record (HER) data, were used in this study. They found that the normal lung volume, normal lung percentage (NLperc), muscle volume, fat volume, muscle-fat ratio, age, sex, and lesion percentage were the most important variables for predicting mortality and ICU hospitalization. The area under the ROC curve was approximately 0.77 (42). Other studies have also explored the use of deep learning algorithms in analyzing body composition on CT scans to predict outcomes in COVID-19 patients. In this context, Zhang et al. (as cited by Nachit et al.) used a deep learning algorithm to analyze body composition on CT scans and found that myosteatosis was a key predictor of mortality in asymptomatic adults (43). These findings suggest that deep learning algorithms can be useful in predicting outcomes in COVID-19 patients based on body composition analysis. Further research is needed to establish the clinical utility of these algorithms in COVID-19 management.
Machine learning algorithms have been used in many studies to predict COVID-19 mortality. Some studies have used only clinical features, while others have incorporated radiological features as well. The selection of ML algorithms was based on related studies in the field and the quality of the selected dataset. The most commonly used algorithms were SVM, MLP, RF, KNN, and kStar. The performance of the models was evaluated using metrics derived from the confusion matrix, such as AUC and MCC. Important predictors for COVID-19 patient mortality included lymphocyte count, CRP, age, mean lung density, lung tissue percentage, RBC, D-Dimer, and emphysema. The AUC of the models ranged from 0.74 to 0.96. Some studies also used deep learning techniques and EHR data to predict mortality or hospitalization in COVID-19 patients.
In most studies, only the ROC curve, which is a function of the accuracy of predictions, is reported, typically yielding good results. However, in the present study, the agreement of the 4 cells in the contingency table was calculated using MCC. This showed that, although the model may perform well in predicting patient improvement, it may not perform as well in predicting patient mortality, which is the primary concern. For example, in the Gong study, it was shown that all confusion matrix indices focus solely on false positives, while only the MCC Index takes into account both false positives and false negatives (44).
5.1. Conclusions
The main limitations of our study include the possibility that our analysis may not fully account for confounding factors that could influence patient outcomes, such as variations in treatment protocols or differences in healthcare access across different populations. We suggest that simulation studies should be used to enhance understanding and create appropriate indices for machine learning methods, which can be selected based on the type of data. The three variables with the greatest impact on predicting mortality in COVID-19 patients were related to laboratory results, with age being the next most significant variable. Therefore, we recommend that, due to cost, HRCT should only be performed if risk factors are observed in laboratory results, and if necessary, HRCT should be performed promptly.