1. Background
Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) strains are opportunistic pathogens and have been associated with various infections, such as urinary tract infections, septicemia, respiratory tract infections, wound infections and diarrhea (1-4). Extended-spectrum β-lactamases (ESBLs) have emerged worldwide, as a significant cause of community and healthcare-associated infections (5-8). The ESBLs consist of a heterogeneous group of plasmid-mediated bacterial enzymes that confer significant resistance to oxyimino-cephalosporin and monobactam antimicrobials (7, 9-12). The emergence of ESBL-producing bacteria, particularly E. coli and K. pneumoniae, is now a critical concern for the development of therapies against bacterial infection (7, 12-15). Various authors have reported the prevalence of ESBLs to be in the range of 6 ‒ 88% in various hospitals, especially for K. pneumoniae and E. coli (6, 8, 9, 16, 17). The production of β-lactamases is the most common mechanism responsible for resistance to β-lactams among clinical isolates of the Enterobacteriaceae family (4, 7, 11, 13, 16, 18, 19). Systemic infections with ESBL producing Enterobacteriaceae are associated with severe adverse clinical outcomes (9, 20, 21). It is therefore essential for a diagnostic microbiology laboratory to employ updated methods for the detection of ESBL producing strains, taking into account the local epidemiology of ESBL genotypes and their various expression profiles. The ESBL genes are mostly plasmid encoded (9, 11, 22), and the majority ESBLs can be divided into three genotypes: TEM, SHV, and CTX-M (12, 16, 17, 22, 23). During the 1990s, ESBL-producing organisms were described mainly as members of the TEM and SHV-β-lactamase families, in E. coli and K. pneumoniae causing nosocomial outbreaks (6, 20, 22). In the late 1990s, a novel type of ESBL, the CTX-M enzyme, emerged worldwide. The CTX-M types can be divided into five groups, based on their amino acid identities: (CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25) (14, 17, 20, 24). There are multiple studies have reported the prevalence of ESBLs around the world. However, no information is available on the prevalence and types of ESBLs in Syria.
2. Objectives
This study aimed to determine the prevalence of ESBL production among clinical isolates of E. coli and K. pneumoniae and their antibiotic resistant patterns, at the Aleppo University Hospitals. Also, the study investigated the most common ESBL genes, namely: CTX-M, SHV, and TEM, responsible for the ESBL phenomenon among E. coli and K. pneumoniae isolates.
3. Materials and Methods
3.1. Study Location
This study was performed at three University Hospitals in Aleppo city center, Syria, between October 2010 and March 2012. These are the major teaching hospitals in the northern part of Syria.
3.2. Bacterial Isolates
A total of 199 non-duplicate clinical isolates of E. coli (159 isolates) and K. pneumoniae (40 isolates) were collected from 199 patients, with different sites of infections. These isolates were identified using traditional bacteriological methods and biochemical testing, with an API 32 E system (bioMerieux SA, Marcy l’Etoile, France) (25), according to the manufacturer’s recommendations. The isolates were stored at −80°C in MicroBank cryovials containing 20% glycerol (Pro-Lab Diagnostics, Round Rock, TX, USA).
3.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed using the disc diffusion method, recommended by the Clinical and Laboratory Standards Institute (CLSI, 2010) (26), with discs from Abtek Biologicals Ltd. (Liverpool, UK). The results were interpreted according to the current guidelines of the (CLSI, 2010) (26). The following antibiotics were tested: Cefotaxime 30 μg, Ceftazidime 30 μg, Cefteriaxone 30 μg, Chloramphenicol 30 μg, Amoxycillin-clavulanic acid 30/10μg, Piperacillin-tazobactam 100/10 μg, Trimethoprim-sulfamethoxazol 1.25/23.75 μg, Gentamycin 10 μg, Tetracycline 30 μg, Cefepime 30 μg, Imipenem 10 μg, Meropenem 10 μg, Nalidixic acid 30μg, Ofloxacin 25 μg, Ciprofloxacin 5 μg and Amikacin 30 μg.
3.4. Detection and Confirmation of ESBL Production
E. coli and K. pneumoniae isolates were screened for ESBL production using double disk synergy test (27). The ESBL production was confirmed by the E-test method (27), (bioMerieux, Marcy l’Etoile, France). The K. pneumoniae ATCC 700603 was used as a positive control (presence of blaSHVgene) and E. coli ATCC 25922 was used as a negative control (27), (obtained from American Type Culture Collection, Rockville, MD, USA), for both phenotypic methods and for PCR experiments.
3.5. Extraction of Plasmids
Plasmid DNA was isolated from bacterial cells by alkaline lysis method, using the QIAprep Spin Miniprep Kit, which was fully automated on the QIAcube device (Qiagen GmbH, Hilden, Germany), as described by the manufacturer’s instructions. The purified plasmid was stored at -20°C.
3.6. Detection of BlaCTX-M, BlaSHV, and BlaTEM Genes
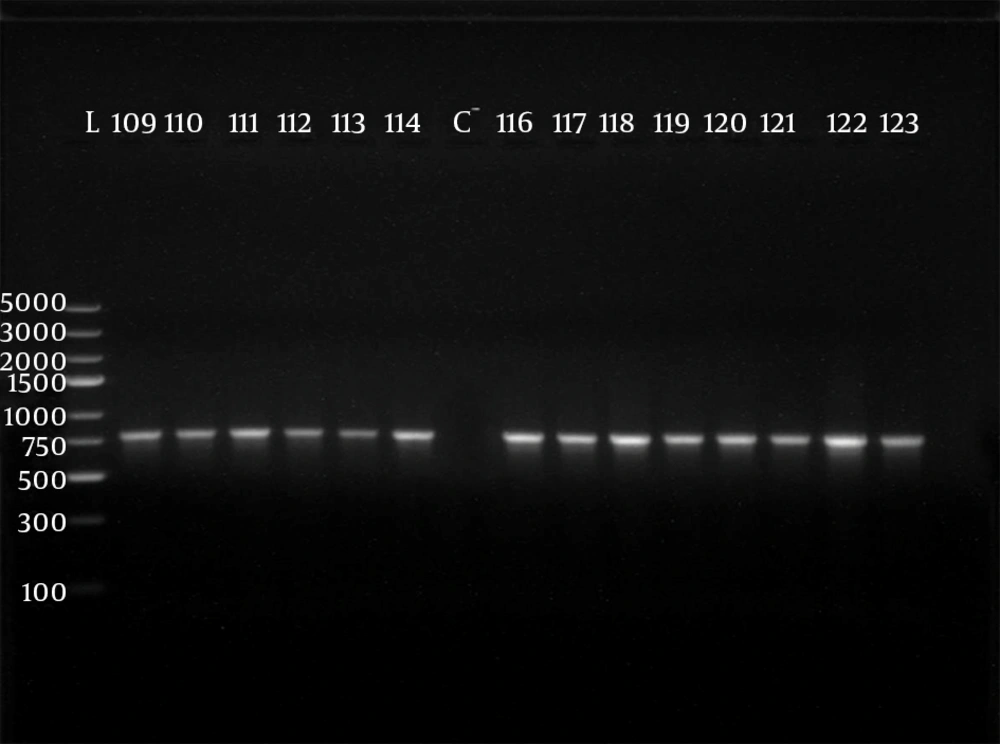
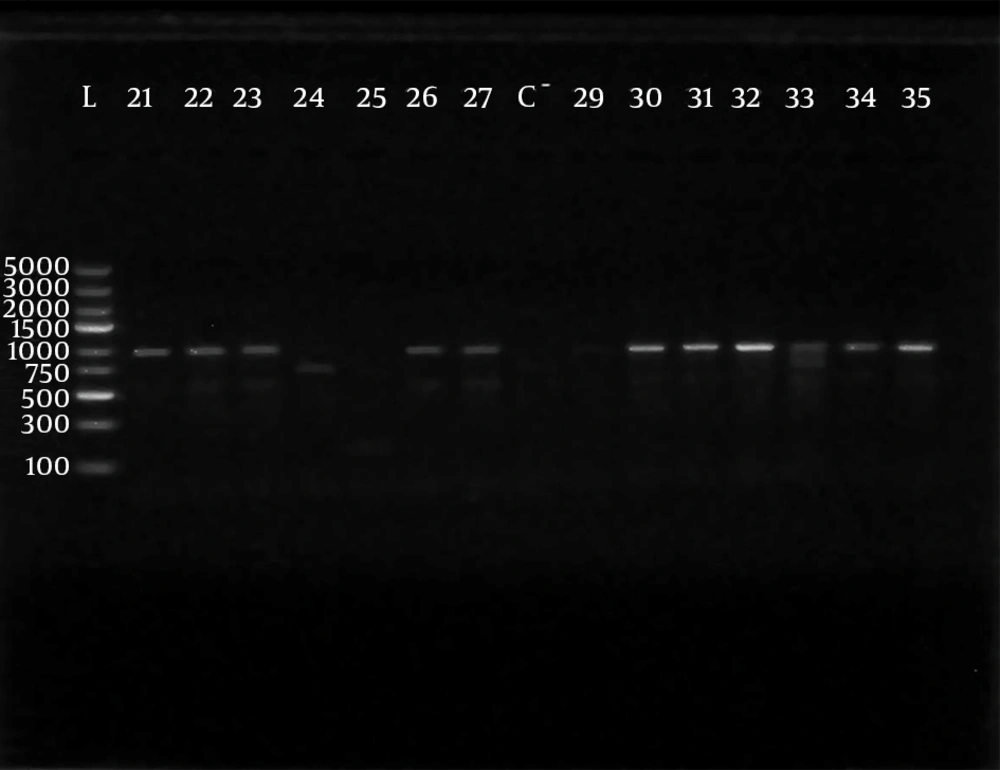
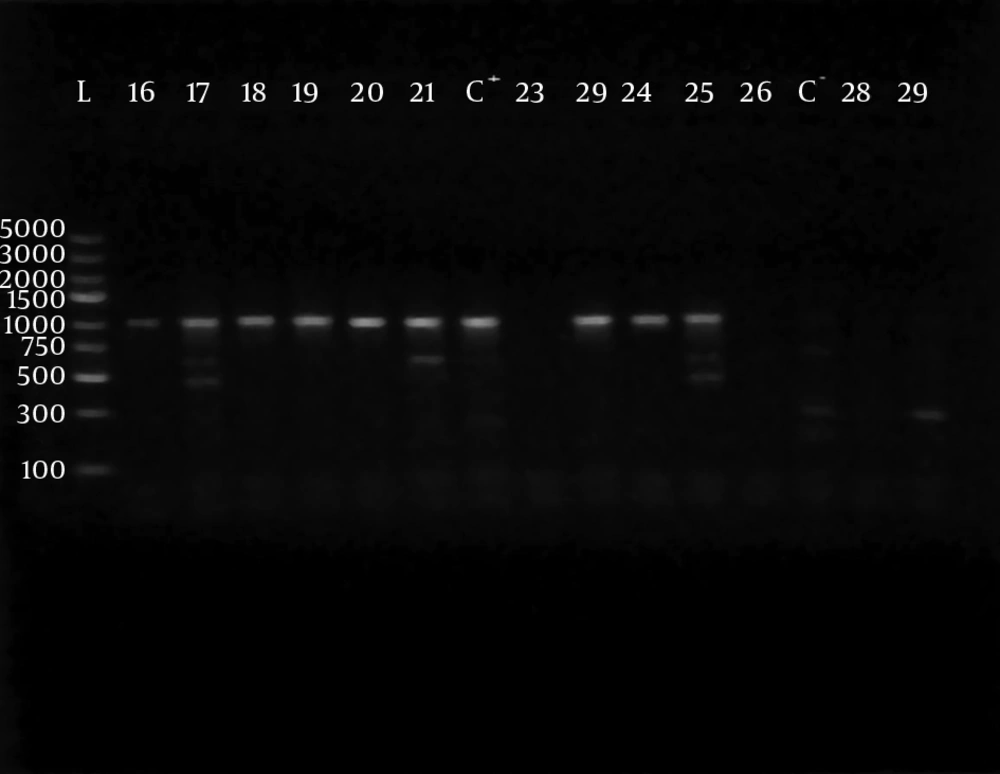
Polymerase chain reaction (PCR) was performed using 5 sets of previously published primers (28, 29) to amplify type-specific ESBL genes, including blaCTX-M-1, blaCTX-M-2, blaCTX-M-9, blaTEM and blaSHV. These primers and are listed in Table 1. The amplified products were analyzed after electrophoresis in 1% agarose gel at 120 V, for 35 minutes. The gel was visualized by staining with ethidium bromide (0.5 μg/mL) in a dark room. A 100 ‒ 5000 bp ladder molecular weight marker (Fermentas, Vilnius, Lithuania) was used. The images of ethidium bromide stained DNA bands were documented using the NiseUV gel documentation system (DAIHAN Scientific, Jijeong-myeon, Wonju-si, Gangwon-do, Korea).
3.7. Statistical Methods
We used the statistical program SPSS version 19 (SPSS Inc., Chicago, IL, USA). Differences between proportions were analyzed using Chi-square test (χ2). All differences, in which the probability of the null hypothesis was P ≤ 0.05, were considered significant.
4. Results
During the study period, 199 consecutive clinical isolates were isolated. Of these, 159 (80%) were identified as E. coli and 40 (20%) as K. pneumoniae. Primary antimicrobial susceptibility test showed that the strongest resistance (exceeding 50%) was to third–generation cephalosporins, nalidixic acid and tremithoprim-sulfamethoxazol, while the majority of these isolates were susceptible to imipenem, meropenem and amikacin. Of 199 isolates, 127 (63.82%) were ESBL producers. Among the ESBL producers 100/159 (62.89%) were E. coli and 27/40 (67.5%) were K. pneumoniae. All ESBL isolates had a minimum inhibitory concentration (MIC) > 16 µg/mL to cefotaxime, more than 96% had MIC > 2 µg/mL and 43% of them had MIC > 32 µg/mL to ceftazidime. The ESBL-producing E. coli and K. pneumoniae isolates had a significantly higher resistance to other groups of antibiotics beyond beta-lactams (Table 2).
The phenotypically identified ESBL-producing E. coli and K. pneumoniae were subjected to PCR using blaCTX-M-1, blaTEM, blaSHV, blaCTX-M-9and blaCTX-M-2specific primers. Of the 98 phenotypically ESBL-producing E. coli isolates (there were two isolates that died during frozen storing), 88/98 (89.80%) were positive for ESBL genes and 10/98 isolates (10.20%) were negative. For phenotypically ESBL-producing K. pneumoniae, all isolates 27 (100%) were positive for ESBL genes. The detected blaCTX-M-1, blaTEM, blaSHV and blaCTX-M-9genes were present alone or in combination with each other. The CTX-M-1 type was the most prevalent among both E. coli and K. pneumoniae isolates, followed by the SHV and TEM types, respectively, while blaCTX-M-2 type were not detected (Table 3). The results of PCR-products electrophoresis showed genomic patterns related to blaCTX-M-1 (780 bp), blaSHV (1007 bp), blaTEM (971 bp) and blaCTX-M-9 (857 bp), (Figure 1 - 3). Seventy three percent of the isolates had two or more ESBL genes. All ESBL K. pneumoniae tested had at least two ESBL genes, while 57 tested isolates (65%) of E. coli had at least two ESBL genes, (Tables 4 and 5).
| Primer Name | Primer Sequence (5´–3´) | Amplicon Size, bp | Ref |
|---|---|---|---|
| CTX-M-1 | 780 | (28) | |
| Forward | CGT CAC GCT GTT GTT AGG AA | ||
| Reverse | ACG GCT TT CTG CCT TAG GTT | ||
| CTX-M-2 | 901 | (28) | |
| Forward | TTA ATG ATG ACT CAG AGC ATT C | ||
| Reverse | GAT ACC TCG CTC CAT TTA TTG | ||
| CTX-M-9 | 857 | (29) | |
| Forward | GTG ACA AAG AGA GTG CAA CGG | ||
| Reverse | ATG ATT CTC GCC GCT GAA GCC | ||
| TEM | 971 | (28) | |
| Forward | TCG GGG AAA TGT GCG | ||
| Reverse | TGC TTA ATC AGT GAG GCA CC | ||
| SHV | 1007 | (28) | |
| Forward | GCC GGG TTA TTC TTA TTT GTC GC | ||
| Reverse | ATG CCG CCG CCA GTC A |
| Antibiotics | ESBL (n = 127) | Non ESBL (n = 72) | P Value |
|---|---|---|---|
| Gentamycin | 69 (54.33) | 5 (6.94) | < 0.0001 |
| Tetracycline | 84 (66.14) | 27 (37.5) | < 0.0001 |
| Nalidixic Acid | 92 (72.44) | 25 (34.72) | < 0.0001 |
| Ciprofloxacin | 67 (52.76) | 8 (11.11) | < 0.0001 |
| Tremithoprim-Sulfamethoxazol | 92 (72.44) | 34 (47.22) | < 0.0001 |
a Data are presented No.(%), n = number of isolates.
bAbbreviation: ESBL, extended spectrum β-lactamases.
| Genotype of Enzymes | Bacterial Species | ||
|---|---|---|---|
| K. pneumoniae (n = 27) | E. coli (n = 88) | Total (n = 115) | |
| CTX-M-1 | 27 (100) | 67 (76.14) | 94 (81.74) |
| SHV | 25 (92.59) | 51 (57.95) | 76 (66.09) |
| TEM | 16 (59.59) | 30 (34.09) | 46 (40) |
| CTX-M-9 | 0 (0) | 5 (5.68) | 5 (4.35) |
| CTX-M-2 | 0 (0) | 0 (0) | 0 (0) |
a Data are presented as No.(%), n = number of isolates.
| Genotypes | K. pneumoniae (n = 27) |
|---|---|
| CTX-M-1 + SHV + TEM | 14 (51.85) |
| CTX-M-1 + SHV | 11 (40.74) |
| CTX-M1 + TEM | 2 (7.41) |
| Total | 27 (100) |
a Data are presented as No.(%), n = number of isolates.
| Genotypes | E. coli (n = 88) |
|---|---|
| Two or Three Combination Genes | |
| CTX-M-1 + SHV | 27 (30.68) |
| CTX-M-1 + TEM | 17 (19.32) |
| CTX-M-1 + SHV + TEM | 8 (9.09) |
| SHV + CTX-M-9 | 3 (3.41) |
| SHV + TEM | 2 (2.27) |
| Total | 57 (64.78) |
| One Genotype | |
| CTX-M-1 | 15 (17.05) |
| SHV | 11 (12.5) |
| TEM | 3 (3.41) |
| CTX-M-9 | 2 (2.27) |
| Total | 31 (35.22) |
a Data are presented as No.(%), N = number of isolates.
5. Discussion
Knowledge on local antimicrobial resistance trends among clinical isolates is important for evidence based recommendations in the empirical antibiotic treatment of infections. The current study described the antimicrobial resistance rates including detection of ESBL among E. coli and K. pneumoniae isolates, which are the predominant ESBL producers. In the current study, the antimicrobial resistance rates of both E. coli and K. pneumoniae isolates was high to first line antimicrobial agents, such as cotrimoxazole, amoxicillin/clavulanic acid, nalidixic acid, ceftriaxone and ciprofloxacin. Our result is consistent with multiple studies (8, 14, 18, 24, 30-33). The observation may be due to the wide use of these drugs empirically, because they are relatively cheap and also, by being oral antibiotics, they are easy to administer. In the present study, the carbapenems (imipenem and meropenem) and amikacin were very active antimicrobial agents against the ESBL and non-ESBL-producing E. coli and K. pneumoniae isolates. This is in agreement with other studies (3, 9, 10, 13, 18, 23, 31). During the past decade, ESBL producing Gram-negative bacilli, especially E. coli and K. pneumonia have emerged as serious pathogens, both in hospital and community acquired infections, worldwide. The occurrence of ESBL among clinical isolates greatly varies worldwide and geographically, and is rapidly changing over time (14, 24, 33). In the present study, ESBL phenotypes were found to be positive in 127 isolates (63.82%). We demonstrated a high prevalence of ESBL production by E. coli (62.89%) and K. pneumoniae (67.5%) isolates, at Aleppo Univesity Hospitals, Aleppo, Siria. Similar results were reported in Jordan (10) (50.8%), Sudan (11) (59.6%), Iran (34) (59.2%), India (17) (63.6%) of E. coli and (66.7%) of K. pneumoniae and in Turkey (35), (39.4%) of E. coli and (60.6%) of K. pneumoniae. However, our result is higher compared to other reports from Germany (36) (1.6%) of enterobacteriaceae, Switzerland (20) (0.7%) of enterobacteriaceae and Saudia Arabia (6), where (20.4%) of E. coli were ESBL-producing. On the other hand, other studies in China, Nigeria and India reported high percentages of ESBL-producing isolates, with 94.1% E. coli (4) and (52.4%) K. pneumoniae, in Nigerria (12), (80%) of E. coli and K. pneumoniae and in India (77%) of E. coli and K. pneumoniae were ESBL-producing (37). The high prevalence of ESBL-producing isolates described in this study was probably due to the large amount of third-generation cephalosporins consumed by patients.
Compared with ESBL-negative isolates, ESBL-positive E. coli and K. pneumoniae isolates showed a significantly high rate of resistance to non-β-lactam antibiotics, such as cotrimoxazole, nalidixic acid, tetracycline, gentamycin and ciprofloxacin. This association between ESBL production and decreased susceptibility to non-β-lactams is in line with the findings of previous investigations, (8, 14, 20, 24, 32). This is probably because the ESBL genes are located on a plasmid that can be transferred from one organism to another, rather easily, and can incorporate genetic material coding for resistance to other antimicrobial classes. Our results demonstrated that all ESBL isolates had MIC > 16 μg/mL to cefotaxime and more than 90% had MIC > 2 µg/mL to ceftazidime and 43% had MIC > 32 µg/mL to ceftazidime. Similar results were reported in various studies (19, 31, 32).
In the present study, genotypic survey on 125 confirmed ESBL phenotype strains (there were two isolates that died during frozen store) by PCR revealed that115 isolates (92%) were positive genotypes for at least one of the studied genes. The negative amplification in the remaining isolates may be due to the presence of uncommon of other ESBL genes, which we did not explore further. Of all phenotypically identified ESBL-producing K. pneumonia, 27 isolates (100%) were positive genotypes, compared with 88 isolates of E. coli, 89.80% were positive. The PCR results showed that among ESBL gene families, blaCTX-M-1was the most prevalent in both K. pneumoniae and E. coli ESBL-producing isolates, 100% and 76.14%, respectively. A very high prevalence of blaSHV (92.59%) and (57.95%) was also detected among K. pneumoniae and E. coli isolates, respectively. These results are in agreement with other studies, such as in Saudia Arabia (6), which reported that, 96.8% of CTX-M types in, E. coli belonged to CTX-M-1 group and 3.23% belonged to CTX-M-9 group; the study in Sudan (11) showed that CTX-M gene was found in 71.4% of E. coli and 86.4% of K. pneumonia and in Switzerland (20), mentioned that CTX-M genes were the most prevalent and most of them belong to the CTX-M-1 group. However, the Brazilian study found that the highest prevalence ESBL genes were aTEM followed by CTX-M type.
In conclusion, the results of this study suggest the importance of ESBL-producing E. coli and K. pneumoniae, as a cause of infections in the Aleppo University Hospitals, Aleppo, Siria. The high prevalence of multidrug-resistant organisms should be taken into account when choosing therapeutic agents, while the continuous local monitoring of resistance patterns is necessary to adequately select an empirical antimicrobial therapy. Further studies, aimed at unraveling the molecular mechanisms of resistance, will provide a better understanding of the epidemiology associated with ESBL-producing species of Enterobacteriaceae.