1. Background
Streptococcus pneumoniae is an important bacterial pathogen in children and older adults (1). The S. pneumoniae is a leading cause of bacterial meningitis, pneumonia, bacteremia, and acute otitis media. It is associated with the highest case fatality rate in patients with pneumonia and meningitis (2). In the recent years, penicillin nonsusceptible and multiple antibiotic resistance clinical isolates have emerged in many parts of the world (3-5). Alterations in the penicillin-binding proteins lead to a decrease in the bacteria’s affinity for the antibiotics, and thus decrease the susceptibility to the antibiotics (6). Therefore, increasing resistance rate may cause serious problems in the treatment of pneumococcal infections (6).
Antibiotics such as cephalosporins and vancomycin serve as alternative therapeutic agents in infections with clinical isolates resistance to other antibiotics such as penicillin (7). In addition, emerging resistance to beta-lactam, resistance to macrolides, tetracycline and trimethoprim-sulfamethoxazole were reported (8). Increase in the use of vancomycin for invasive pneumococcal infections may help develop resistance to this important drug; moreover, vancomycin tolerated strains of S. pneumoniae have been characterized in the recent studies. To date, vancomycin-resistant strains of pneumococci have not been seen, but strains of S pneumoniae tolerant to vancomycin have been reported (9).
2. Objectives
In the present study, we aimed at determining the phenotypic and genotypic properties of vancomycin nonsusceptible S. pneumoniae isolates obtained from clinical isolates in Tehran, Iran.
3. Methods
3.1. Bacterial Isolation and Identification
This was a cross- sectional descriptive study conducted during November 2014 and March 2016 on 5 pneumococcal isolates (isolates selected from a total of 73 S. pneumoniae and collected during the study period), which were obtained from different clinical specimens in 2 hospitals (Milad and Sina) in Tehran, Iran. The isolates were transported to department of microbiology affiliated to Shahid Beheshti University of Medical Sciences for further analysis. All suspected S. pneumoniae isolates were streaked for single colonies on blood agar, supplemented with 5% sheep’s blood (Merck, Germany, Lot No. VM1287860091). All plates were incubated for 24 to 48 hours at 37°C in candle jar. All isolates were identified as S. pneumoniae based on colonial morphology, hemolysis, gram staining, bile solubility, and susceptibility to optochin disc (1 μg, Mast, UK, Lot No.303005). S. pneumoniae ATCC 49619 was used for quality control. Identification of the isolates as S. pneumoniae was confirmed by PCR for the cpsA gene (coding for capsular polysaccharide biosynthesis) by species-specific primers (8). A medium containing skim milk, tryptone, glucose, and glycerin (STGG, Merck, Germany, Lot No. VL101263934) was used for the storage of bacteria at -70°C.
3.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility tests of pneumococcal isolates were performed by the broth microdilution method according to guidelines of the clinical and laboratory standards institute (CLSI) (10) for vancomycin, penicillin, cefotaxime, ceftriaxone, erythromycin, trimethoprim-sulfamethoxazol, ofloxacin, and meropenem. The bacterial isolates were incubated overnight on 5% sheep blood agar plates. Inoculants were prepared from the bacterial colonies that formed on the 5% sheep blood agar plates, and then the colonies were suspended in 0.9% saline with a turbidity equivalent of 0.5 McFarland units. The final inoculum density was of 5 × 105 CFU/mL in each test tube. Antimicrobials agents had been diluted in cation-adjusted Mueller-Hinton broth (BD; USA), with 5% lysed horse blood. The following concentrations for the antimicrobials were used: 32 µg/mL, 16 µg/mL, 8 µg/mL, 4 µg/mL, 2 µg/mL, 1 µg/mL, 0.5 µg/mL, 0.25 µg/mL, 0.12 µg/mL, and 0.06 µg/mL. The microplates were incubated at 35º C in ambient air for 20 to 24 hours.
3.3. DNA Extraction and PCR Assay
Bacterial DNA extraction was done by high pure PCR template preparation kit (Roche, Germany). The extracted DNA suspension was kept frozen at -70°C until further use. The polymerase chain reaction (PCR) was performed to detect vanA gene, which was described previously (9).
3.4. Statistical Analysis
Analysis was performed using SPSSTM software, version 21.0 (IBM Corp., USA). The results are presented as descriptive statistics in relative frequency. Values were expressed as the mean ± standard deviation (continuous variables) or percentages of the group (categorical variables).
4. Results
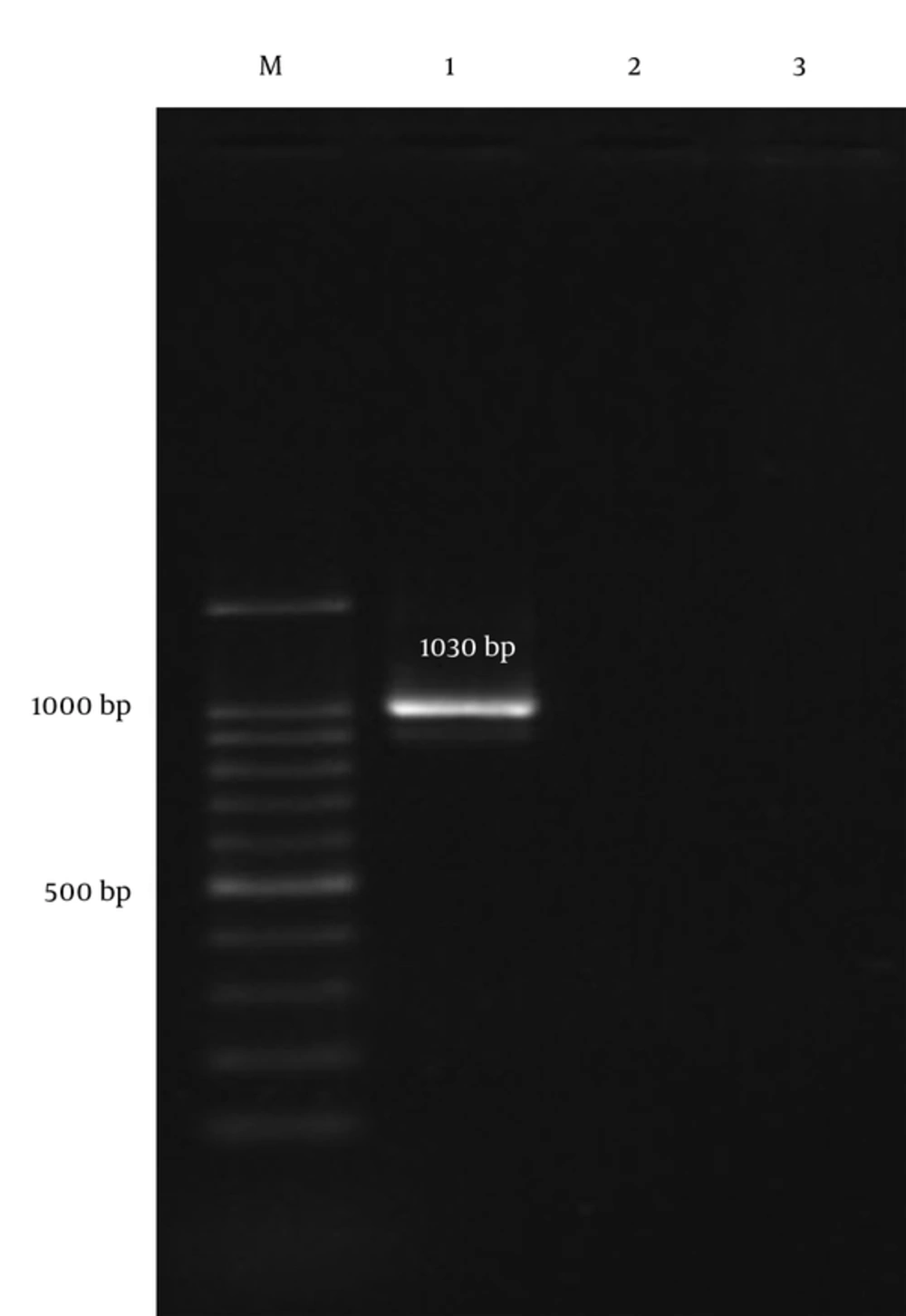
During the study period, 5 S. pneumoniae clinical isolates, which were nonsusceptible to vancomycin, were identified. Table 1 demonstrates the full antibiotic susceptibility pattern and clinical source of pneumococcal isolates. Vancomycin minimum inhibitory concentrations (MIC) for tested isolates ranged from 2 µg/mL to 16 µg/mL. In total, 80%, 60%, and 40% of the isolates were susceptible to ofloxacin, penicillin, and trimethoprim-sulfamethoxazol, respectively. The only penicillin resistant isolate was susceptible to trimethoprim-sulfamethoxazol and resistant to other tested antibiotics. Moreover, all the isolates were resistant to cefotaxime and erythromycin. Finally, our findings showed that all vancomycin nonsusceptible isolates with MIC ≥ 1 µg/mL were PCR negative for vanA gene (Figure 1).
| Isolates | Gender | Age | Sample | VanMIC, µg/mL | PEN | CTX | CRO | ERY | SXT | OFX | MER |
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. 1 | F | 17 < | BAL | 2 | I | R | R | R | I | S | R |
| No. 2 | F | 17 > | BAL | 2 | S | R | I | R | S | S | R |
| No. 3 | F | 17 > | Eye | 16 | R | R | R | R | S | R | R |
| No. 4 | M | 17 < | Blood | 16 | S | R | R | R | R | S | I |
| No. 5 | M | 17 > | CSF | 2 | S | R | R | R | I | S | I |
Antibiotic Susceptibility Pattern and Characteristics of Five Non-Susceptible Vancomycin S. pneumonia
5. Discussion
Antimicrobial resistant pneumococcal isolates are becoming more common throughout the world, especially in developing countries (11). Although some studies indicated that penicillin and cephalosporin are often included in the initial empirical antibiotics regimen for patients with pneumococcal infections, penicillin and cephalosporin-resistant S. pneumoniae isolate are being isolated with an increasing level and become a serious health problem around the world (12). Insufficient information on distribution of antibiotic resistant pneumococci in our geographical area necessitates a serious attention to estimate burden of resistant isolates. In the present study, we investigated a total of 5 S. pneumoniae clinical isolates, which showed resistance to vancomycin with high level of MIC. Our results demonstrated an alarming rate of vancomycin resistant pneumococci, which might have resulted from the uncontrolled and frequent use of vancomycin and self-medication (13).
In this study, 2 phenotypic (broth microdilution) and genotypic (PCR) methods were used to determine susceptible and resistant isolates to vancomycin, and our results demonstrated that all isolates with MIC ≥ 1 µg/mL were negative for the presence of vanA gene.
In previous Iranian studies, range of resistance to penicillin was 0% to 70% among clinical isolates (1, 14). Compared with other reports in Iran, our results showed only 1 pneumococcal isolate resistant to penicillin (20%), therefore, the rate of resistance to penicillin in our isolates was lower than those reported from other studies (15, 16). Our study also found that both penicillin-resistant and penicillin-susceptible strains showed resistance to erythromycin and cefotaxime, which was similar to reports from Zahedan, Iran (15).
In this study, it was observed that isolate No. 3 was not only resistant to vancomycin, but was also resistant to other antibiotics such as ceftriaxone, cefotaxime, ofloxacin, meropenem, erythromycin, and penicillin. Four of the isolates in the present study remained susceptible to ofloxacin, which probably can be recommended as an alternative treatment option for penicillin and macrolides resistant isolates. However, we found no significant differences between source of infection and antibiotic resistance pattern among the studied isolates.
5.1. Conclusions
In summary, it seems that the increasing use of penicillin and vancomycin in the recent years created an opportunity for the emergence of penicillin and vancomycin-resistant clinical isolates. The increase in antibiotic resistance among S. pneumoniae strains in Iran seems to be alarming, and the treatment of infections should be further analyzed and investigated.