1. Background
Staphylococcus epidermidis normally resides on the skin and mucous surfaces. It is most often associated with hospital-acquired infections particularly when implanted medical devices such as urinary tract catheters are used (1-5). It is also the most prevalent bacterium recovered from immunocompromised patients (6, 7). The point that highlights the pathogenesis of S. epidermidis infection is its high resistance to several classes of antibiotics and its ability to form biofilm (7).
The importance of bacterial biofilms was highlighted by Arciola et al., who pointed out that the formation of bacterial biofilm and inherent resistance to antimicrobial agents and to the patient’s immune system are the causes of many persistent and chronic bacterial infections (8). Bacteria in the biofilm are protected from the host defense system and antibiotics administered for the treatment of infections (9, 10). Biofilms usually result in persistent infections that cannot be easily resolved with standard antibiotic treatments (10) because the removal of the foreign body is often necessary for a cure (11, 12). Thus, the associated infections are difficult to clear, causing increased morbidity and mortality. Factors involved in biofilm-associated resistance include limited penetration, decreased growth rate, cell density, unique cell physiology, persister cells, and altered chemical microenvironment (13).
Biofilm-associated accumulation of bacterial cells that are enclosed in a self-produced matrix exopolysaccharide can easily attach to biotic or abiotic surfaces (3, 14-19). This biofilm structure is made up of extracellular matrix that comprises polysaccharides, proteins, enzymes, DNA, etc. (20, 21). Biofilm development depends on many physical, chemical, and biological factors (20). In staphylococci, the main intracellular adhesion molecule in staphylococci is the polysaccharide intercellular adhesion (PIA). Biofilm formation is regulated by the expression of PIA, also known as poly-N-acetylglucosamine (PNAG) (22). PIA participates in cell-cell adhesion and plays an important role in biofilm formation by CoNS (10, 23). The PIA is encoded by ica (intercellular adhesion) genes that are organized in an operon structure. The operon contains the ica ADBC genes (3, 24). Functional analysis of ica ADBC proteins revealed that proteins icaA, icaD, and icaC are present on the cell membrane. IcaB is secreted in the culture supernatant. During PIA synthesis, icaA displays N-acetylglucosaminyltransferase activity and the co-expression of IcaA and IcaD genes increases the transferase activity. The combination of icaA and icaD can produce N-acetylglucosamine oligomers in a maximal length of 20 residues. IcaB, a deacetylase, has shown sequence similarity to the Rhizobium NodB protein. IcaC, a transmembrane protein, may facilitate the translocation of the growing polysaccharide to the cell surface (3, 8, 20). In addition, the ica R gene encodes a transcriptional repressor, which downregulates ica operon expression related to environmental factors in S. epidermidis (20, 25-27).
The production of biofilm is analyzed both by the phenotypic methods, such as the microtiter plate (MTP) assay devised by Christensen et al. and the Congo red agar (CRA) plate test as described by Freeman et al., and by the molecular detection of the ica locus (28-31).
2. Objectives
The aim of the present study was to investigate biofilm production in CoNS strains isolated from clinical and healthy individuals using qualitative Congo red agar (CRA test) and quantitative microtiter plate assay (MTP). The presence of icaA genes was determined by polymerase chain reaction (PCR).
3. Methods
3.1. Bacterial Isolates and Species Identification
This cross-sectional study was performed in a period of 12 months from June 2016 to June 2017 at Hashemi Nejad Hospital in Tehran, Iran. In total, 50 isolates of S. epidermidis were collected from inpatients. The samples included blood, medical devices, wound, pus, and urine. All specimens were cultured on blood agar plates, incubated at 37˚C for 24 hours, and assessed by standard biochemical and microbiological tests to identify S. epidermidis.
3.2. Antibiotic Susceptibility Testing
For the antibiotic susceptibility testing, isolates were suspended in nutrient broth and the suspension was adjusted to a turbidity equivalent to a 0.5-McFarland standard, corresponding to 108 CFU/mL. The antibiotic susceptibility test was performed using the disk diffusion method. Muller Hinton plates were incubated at 35°C and read after 18 hours of incubation. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. Eleven antibiotic discs were tested: amikacin (30 μg), kanamycin (15 μg), minocycline (30 μg), vancomycin (30 μg), clindamycin (2 μg), cotrimoxazole (25 μg), nitrofurantoin (300 µg), erythromycin (15 μg), Synercid (15 μg), penicillin (10 μg), and rifampin (5 µg).
3.3. Detection of Biofilm Formation
3.3.1. Congo Red Agar (CRA) Method
The Congo red test was performed on brain heart infusion agar (BHI agar, 37 g/L), containing glucose (36 g/100 mL), and Congo red stain (0.8 g/100 mL). The plates were incubated aerobically for 24 to 48 hours at 37°C. Biofilm producers formed black colonies on CRA whereas non-producers formed red colonies. The Congo red dye directly interacts with certain polysaccharides to form colored complexes.
3.3.2. Microtiter Plate (MTP) Method
This quantitative test described by Christensen et al. is considered the gold standard method for biofilm detection. Organisms isolated from fresh agar plates were inoculated in 10 mL of trypticase soy broth with 1% glucose. Cultures turbidity was adjusted to a 0.5-McFarland standard. Individual wells of sterile 96-well flat bottom polystyrene tissue culture plates were treated with 200 µL of the bacterial suspension. Negative control wells contained sterile broth. Plates were incubated at 37°C for 24 hours. After incubation, the content of each well was removed by gentle tapping. The wells were washed four times with 0.2 mL of phosphate-buffered saline (pH 7.2). The washing removed free-floating bacteria. The adherent bacteria were fixed in 2% sodium acetate, dried at 65°C for 1 hour, and stained with 1% crystal violet for 15 minutes. Excess stain was removed by gently washing the plates twice with distilled water. Finally, 100 µL of an alcohol solution containing 70% ethanol and 10% isopropyl alcohol was added to each well, and optical density was measured at 570 nm. All tests were performed in triplicate. Biofilm production was considered as strongly adherent (optical density of higher than 0.3), moderate adherent (optical density of 0.2 - 0.3), weakly adherent (optical density of 0.1 - 0.2), and non-adherent (optical density of lower than 0.1).
3.3.3. Polymerase Chain Reaction (PCR)
Cellular DNA was acquired from S. epidermidis colonies grown overnight on blood agar plates by the High-Pure PCR Template Preparation Kit (Roche Co., Germany) according to the manufacturer’s instruction. Species-specific primers were used for detection of the icaA gene. The primer sequences for icaA were: forward (5’-GacCTCgAAgTC AATAgAggT-3’) and reverse (5’-CCCAgTATAACgTTggATACC-3’). The two primers amplified an 814-bp region. A total volume of 25 µL PCR reaction mixture containing 1 µL of template DNA, 12.5 μL of Master mix 2X (Amplicon, Denmark), and 2 µL of each primer was utilized. Reactions were performed in a thermal gradient cycler (Eppendorf Co., Germany) with the following thermal cycling profile: an initial denaturation at 94°C for 5 minutes, followed by 30 cycles of amplification (denaturation at 94°C for 1 minute, annealing at 59°C for 1 minute, and extension at 72°C for 1 minute) and a final extension period of 5 minutes at 72°C. Amplification products were evaluated by electrophoresis on the 1% agarose gel (Tris-acetate buffer) and visualized by safe stain (Cinna Gen).
4. Results
4.1. Isolates and Identification of Coagulase-Negative Staphylococci
In the current study, 50 isolates of S. epidermidis were collected from various clinical specimens. The distribution of isolates was as follows: Blood (n = 20, 40%), urine (n = 4, 8%), pus (n = 8, 16%), catheter (n = 10, 20%), and wound (n = 8, 16%).
4.2. Congo Red Agar
The phenotypic production of slime by all strains under study was assessed by culture on CRA plates.
Of the 50 clinical samples, 34 (68%) isolates were biofilm producers. Biofilm-producing bacteria were isolated from blood (22%), medical devices (14%), pus (10%), urine (8%), and wound (14%).
4.3. Microtiter Plate Method (MTP)
Of the 50 clinical samples, 38 (76%) isolates were positive for biofilm formation in the MTP test. Biofilm-producing bacteria were isolated from blood (28%), medical devices (14%), pus (10%), urine (8%), and wound (16%).
4.4. PCR for IcaA Gene
Of the 50 clinical samples, 36 (72%) isolates were positive for the icaA operon. Biofilm-producing bacteria were isolated from blood (26%), medical devices (14%), pus (10%), urine (8%), and wound (14%).
4.5. Correlation Between the Presence of the Ica Operon, Biofilm Production, and the MTP Results
Comparison between the results of MTP test and the results obtained by PCR revealed that 68% of the isolates were concomitant positive. Thee biofilm formation comparison between the results of MTP test and the results obtained by CRA revealed that 68% of the isolates were concomitant positive. Comparison between the results of the CRA test and the results obtained by PCR revealed that 58% of the isolates were concomitant positive.
4.6. Antibiotic Susceptibility Testing
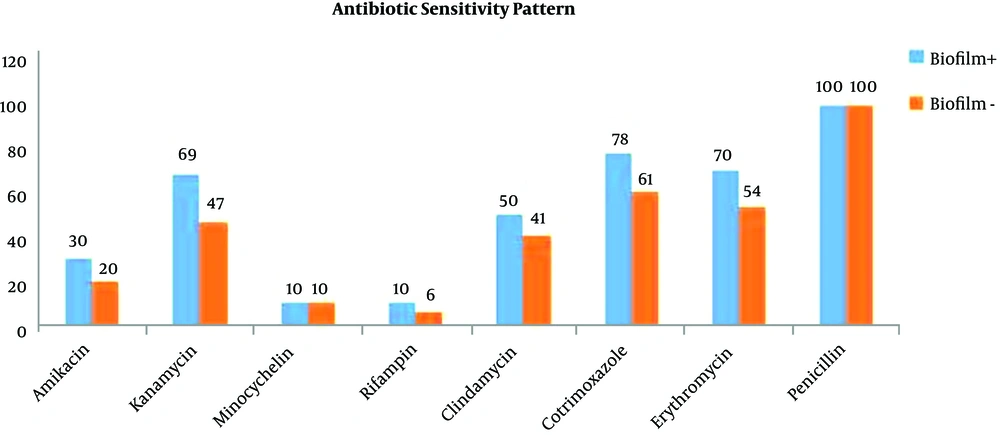
Antibiotic susceptibility tests were performed by the Kirby-Bauer disk diffusion method following the CLSI guidelines. The international reference strains were used as controls including S. epidermidis ATCC 29213 and S. epidermidis ATTC25923 as standard positive and negative control strains, respectively. The results of the study indicated that all of the isolates were sensitive to nitrofurantoin, vancomycin, and Synercid. The most resistance was noted toward penicillin. The percentages of resistance to each antibiotic are presented in Table 1 and Figure 1.
| Antimicrobial Agents | Biofilms Producers (%) | Non-Biofilm Producers (%) |
|---|---|---|
| Vancomycin | 0 | 0 |
| Synercid | 0 | 0 |
| Nitrofurantoin | 0 | 0 |
| Penicillin | 100 | 100 |
| Amikacin | 30 | 20 |
| Kanamycin | 69 | 47 |
| Minocycline | 10 | 10 |
| Rifampin | 10 | 6 |
| Clindamycin | 50 | 41 |
| Cotrimoxazole | 78 | 61 |
| Erythromycin | 70 | 54 |
5. Discussion
Coagulase-negative staphylococci are implicated in hospital infections and they can infect a wide variety of prosthetic medical devices. The most important virulence factor of these bacteria is their ability to stick to hard surfaces and form biofilms (32). The ability of staphylococci to form biofilms helps the bacteria to resist the host immune response and antimicrobial agents (22).
It has been noticed in several studies that Staphylococcus epidermidis is the most frequently isolated species in nosocomial infections and is the most common causative organism found in infections created by implanted medical devices. It makes up a significant part of the normal bacterial flora of the human skin and mucous membranes and it is probably introduced easily as a contaminant during the surgical implantation of polymeric devices. Our study used three widely used methods for testing biofilm formation capability, including growth on CRA, MTP method, and PCR-based detection of ica operon.
The results of the current study indicated that all the isolates were susceptible to vancomycin, nitrofurantoin, and Synercid. The most resistance was noted toward penicillin. This correlated well with the results obtained by Gordon et al. in the USA that reported all isolates were susceptible to vancomycin and 98% of the isolates were resistant to penicillin (33).
Of the 50 samples (isolates), 34 (68%) were biofilm producers in the CRA test. This correlated well with the study by Mendoza et al. in Mexico that reported biofilm detection in 62% of the isolates by the CRA method. It was also in accordance with the study by Mertens and Ghebremedhin in Germany that reported biofilm detection in 64% of the isolates by the CRA method (12, 34). Biofilm-producing bacteria were most isolated from blood (22%), followed by medical devices (14%), pus (10%), urine (8%), and wound (14%).
Using the CRA method, Silva et al. detected biofilm production in only 25% of the CoNS strains isolated from clinical specimens of newborns in the NICUs (10). The CRA test seems to be easier and faster than other methods for measuring biofilms. However, its results are comparable with the results of the molecular method. The same approach was used by Fitzpatrick et al. (10, 35).
Of the 50 samples, 38 (76%) were positive in the MTP test. The results correlated well with those by Oliveira and Cunha Mde in Brazil that detected biofilm formation in 81% of the isolates, Hell et al., in the USA that reported biofilm detection in 63% of the isolates, and Tremblay et al. in Canada that reported biofilm formation in 85% of the isolates by the MTP method (2, 10, 14).
Biofilm-producing bacteria were most isolated from blood (28%), followed by medical devices (14%), pus (10%), urine (8%), and wound (16%). In the study by Mathur et al. 57.8% of staphylococcal clinical isolates displayed a biofilm-positive phenotype and 14.47% and 39.4% exhibited high and moderate biofilm formation, respectively (36); however, in 46% of the isolates, weak or no biofilm formation was detected. In the study by Oliveira et al., among 100 isolates studied, 35 (35%) were classified as weakly adherent and 46 (46%) as strongly adherent (10).
Bose et al. reported that 54.19% of staphylococci were biofilm producers whereas Mathur et al. reported a rate of 53.9% (37). The MTP assay is the most useful and most widely used technique as a standard test for the detection of biofilm formation (37). This method is the most sensitive and most accurate method and has the advantage of being a quantitative tool for comparing the adherence of different strains (10).
In our current study, the PCR method was used to detect the ica operon. Since PCR is a simple technique, it can be an important tool for the identification of ica genes. It is relatively rapid and reliable and requires minimal amounts of DNA. PCR was used as a reference for the phenotypic method based on several studies. In addition, the ica genes are important virulence markers of clinical CoNS isolates since their expression is associated with the production of PIA, the most clearly characterized component of staphylococcal biofilms (22). In this study, of the 50 isolates, 36 (72%) were positive for the icaA operon. This was in accordance with the study by Oliveira and Cunha Mde, who reported that 82% of the isolates carried the ica operon (10).
Comparing the MTP test results with the PCR assay results revealed that 68% of the clinical isolates and 38% of the healthy individuals’ isolates were biofilm-positive. Comparison between the MTP test and the results obtained by CRA revealed that 68% of the isolates were positive for biofilm formation. Comparison between the CRA test and the results obtained by PCR revealed that 58% of the isolates were positive for biofilm formation. In the present study, the MTP test showed the best correlation with the PCR results. Moreover, biofilm-producing strains were more resistant than non-biofilm producers.
5.1. Conclusions
Intercellular adhesion genes (ica) and consequent biofilm production were detected in most CoNS isolates. Therefore, infections caused by biofilm-producing staphylococci could complicate antibiotic therapy. The MTP method showed the best correlation with the PCR results. Antibiotic resistance and biofilm forming potential were more prevalent in clinical isolates. Since the pathogenic determinants of these bacteria are very complex, multifactorial, and dependent on numerous genetic and environmental factors, more accurate molecular studies are needed in this context. Other genes that may contribute to the infection process are also involved in biofilm formation in coagulase-negative staphylococci, which need to be more studied. Additionally, gene regulation rather than the presence or absence of the ica operon may be involved in bacterial virulence.