1. Background
Leishmaniasis includes a group of parasitic diseases caused by species of obligating intra-macrophage protozoan Leishmania parasites (1). These parasites are transmitted to humans and animals by the bite of phlebotomine sand flies (1). Of 20 species of Leishmania, 18 species are zoonotic, creating a wide range of clinical symptoms such as cutaneous, diffuse cutaneous, mucocutaneous (espundia), and visceral (kala-azar) symptoms, as well as post kala-azar dermal leishmaniasis (PKDL) and recidivans (2). More than 90% of cutaneous leishmaniasis (CL) cases occur in America, Mediterranean basin, Middle East, and Central Asia regions. According to the World Health Organization (WHO), more than two-thirds of new CL cases occurred in Afghanistan, Algeria, Brazil, Colombia, Iran, and the Syrian Arab Republic (3). Approximately 0.7 to 1.2 million new cases are reported each year, with 20,000 to 40,000 leishmaniasis deaths annually (4). Many Leishmania infections are either asymptomatic or misdiagnosed (5, 6). In Iran, two species of Leishmania tropica and Leishmania major cause anthroponotic cutaneous leishmaniasis (ACL) and zoonotic CL (ZCL), respectively. Furthermore, visceral leishmaniasis (VL) is caused by L. donovani complex (7). Most ACL cases are reported from northeast (Khorasan-Razavi) and Southeast (Kerman) regions of Iran, while ZCL is distributed in northeast (Khorasan-Shomali), Central (Isfahan), and West (Ilam), as well as Southern provinces (Khouzestan and Bushehr). In Iran, more than 90% of the visceral cases are reported from the northwest and southern provinces, including Fars and Bushehr (7).
In recent decades, new leishmaniasis foci were expanded or emerged in Iran, indicating epidemiological changes in leishmaniasis distribution. Remarkably, these new foci have now become endemic for leishmaniasis, especially in tegumentary form (8-10). Some well-known reasons for this phenomenon include changes in environmental conditions, such as climate change, and human activities such as travel to endemic foci and migration (11). In Iran, nearly 18,000 - 20,000 new cases of CL are reported annually. Because of spontaneous healing of CL ulcers or delayed refer to health centers, the real number of the disease is estimated 4 - 5 times greater than the formal reports (3).
Prevention, control, and treatment of leishmaniasis depend on several factors, including the type of the disease, parasite species, and geographic location. In CL, the disease is diagnosed based on the clinical symptoms and parasitological tests (12). Leishmania spp. are directly identified through morphology studies in microscope slides of the clinical samples. Since several Leishmania spp. can be found in one study, and due to impossible differentiation of the species based on their morphology, direct microscopy lacks the necessary sensitivity and specificity (13). Molecular methods such as polymerase chain reaction (PCR) include good sensitivity and specificity. However, choosing target genes is important based on the study aims (13). Various types of PCR, including polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and nested-PCR, have been used for the diagnosis of Leishmania spp., as well as their species identification in the last decades (14).
Re-emerging infectious diseases include a group of diseases that have previously existed but are rapidly increasing or changing their geographic ranges. During the last decade, Dezful and its suburbs, as the second city of Khuzestan province in southwestern Iran, was an endemic area of CL with a low incidence rate. However, the disease rapidly increased from 372 new cases in 2013 to 1,917 cases in 2015. Thus, CL is now described as a re-emerging parasitic disease in this area. Khuzestan Province is one of the old endemic foci of CL in Iran (15). A long border with Iraq, where no controls exist over leishmaniasis due to war and political problems, has resulted in an increased number of CL cases. A close distance to endemic CL foci such as Bushehr, Ilam, and Isfahan provinces affects new foci in Khuzestan province (16-19). Several studies have been carried out in Khuzestan to molecularly characterize Leishmania spp. in human cutaneous lesions (15).
2. Objectives
Therefore, the aim of the current study was to genotype the most prevalent CL strains in Dezful region using PCR-RFLP and sequencing of the ITS1 gene.
3. Methods
3.1. Study Area
The present study was carried out in Dezful, Southwestern Iran, locating along the Dez River in Central Zagros. Dezful is hot and humid, with a history dating back to Sassanian Empire. Dezful is restricted to Andimeshk and Aligoodarz Counties (Lorestan Province) from the north, Lali District of Masjed Soleiman City and Gotvand District of Shushtar City from the east, and Shush City from the south and west (20).
3.2. Sampling, DNA Extraction, PCR, and PCR-RFLP
Totally, 196 Giemsa-stained slides from the patients referred to Dezful Health Center during 2015 - 2016 were collected. The samples were sent to the Department of Medical Parasitology and Mycology, Tehran University of Medical Sciences, Tehran, Iran, for further studies. The preliminary information of the patients, including age, sex, living area, as well as the number, site, and duration of lesions, were recorded in questionnaires. Presence of amastigotes was checked by preparing smears from exudates of lesions, then fixing in methanol and staining by Giemsa, and studying under light microscope. Patients were chosen according to WHO protocols of 4+ (1 - 10 parasites/fields), 3+ (1 - 10 parasites/10 fields), 2+ (1 - 10 parasites/100 fields), and 1+ (1 - 10 parasites/1000 fields) (5). DNA was extracted from 61 samples using DNA extraction kit according to manufacturer’s instructions (Bioneer, Korea). DNA extracts were stored at -20°C until use. PCR on ITS1 gene was carried out using primer set of LITSR (forward: 5’-CTGGATCATTTTCCGATG-3’) and L5.8S (reverse: 5’-TGATACCACTTATCGCACTT-3’) as previously described (21). PCR amplifications were carried out using a thermal cycler in a 25-µL total volume, containing 2.0 µL 10× PCR buffer, 1.2 µL of dNTP mixture (25 mM), 1.6 µL of MgCL2 (25 mM), 1 U of Taq polymerase, 2 µL of each forward and reverse primers (10 pmol), and 1 µL of DNA template (about 20 ng). Amplification results were compared to the standard control from Iranian reference strains of L. tropica (MHOM/IR/02/Mash10/Accession No. EF653267) and L. major (MRHO/IR/11/GOL-2/Accession No.: JN860745). The following condition was used in thermal cycler (Peqlab, Erlangen Germany): an initial denaturing cycle at 95°C for 5 min, followed by 35 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 45 s. The final extension cycle was carried out at 72°C for 7 min. A fragment of approximately 300 - 350 bp was amplified in PCR (21, 22). The PCR products were visualized using electrophoresis on 1.2% safe-stained agarose gels and ultraviolet (UV) light. In RFLP, 10 µL of the PCR products (concentration 1 µg/µL) was mixed with 2 µL of the 10x enzyme buffer and 1 µL of the restriction HaeIII (10 units/µL) enzyme (Fermentas, Germany) and incubated at 37°C for 10 min. Cutting site of the enzyme included GG CC, producing various patterns based on the Leishmania species. Separation of the digested products was carried out using electrophoresis on 3% agarose gels.
3.3. Sequencing and Phylogenetic Analysis
For genotyping and phylogenetic analysis, six samples were sequenced in Bioneer Co., Korea, using ABI 3730 Sequencer. Sequences were checked manually for ambiguities using BioEdit software V.7.1.3.0. DNA sequences were compared to sequences from GenBank Database using Basic Local Alignment Search tool (BLAST). Phylogenetic trees were constructed using the Neighbor-Joining (NJ) method and Tamura 3-parameter for DNA sequences using MEGA 5 software V.5.0) (23). Bootstrap values were set as 1,000 replicates.
3.4. Ethics
The study protocol was approved by the Ethics Committees of Tehran University of Medical Sciences, Iran (approval no.: 34213), and oral consent was obtained from all participants. No information of the patients was revealed in the study.
4. Results
In this study, a total of 196 CL infected patients verified by microscopy (1000×) were selected (Table 1).
| City | Number of Patients (%) |
|---|---|
| Dezful | 55 (28) |
| Shush | 121 (62) |
| Gotvand | 20 (10) |
| Total | 196 (100) |
Distribution of Samples Collected from Acute CL Patients in Dezful and Its Suburbs During 2015 - 2016
Out of 196 patients, 110 (56.2%) and 86 (43.8%) cases were male and female, respectively (Table 2). The average age of the patients was 32 years old, with a mean number of ulcers of two and a lesion duration of 97 days. In 50.5% of the cases, hands were subjects to lesions and 72.15% of the cases included wet lesions. More than half of the cases (60.1%) had one lesions with a mean duration of 32 days.
| Characteristic | No. (%) |
|---|---|
| Age group, y | |
| ≤ 1 | 18 (9.2) |
| 2 - 7 | 65 (33.1) |
| 8 - 15 | 41 (20.9) |
| 16 - 25 | 21 (10.7) |
| 26 - 35 | 20 (10.2) |
| 36 - 45 | 14 (7.2) |
| 46 - 55 | 10 (5.1) |
| 56 - 65 | 5 (2.6) |
| ≥ 66 | 2 (1) |
| Gender | |
| Male | 110 (56.2) |
| Female | 86 (43.8) |
| Number of lesions | |
| Single form | 118 (60.1) |
| Double form | 34 (17.4) |
| Multiple form | 44 (22.5) |
| Location of lesion | |
| Hand | 102 (50.5) |
| Leg | 28 (13.8) |
| Face and head | 62 (30.7) |
| Other parts the body | 10 (5) |
| Lesion appearance | |
| Dry | 17 (27.9) |
| Wet | 44 (72.1) |
| Parasitemia rate | |
| > 4 | 187 (95.5) |
| ≤ 4 | 9 (4.5) |
Main Characteristics and Types of Clinical Lesions in 196 Microscopically Confirmed CL Patients in Dezful and Its Suburbs During 2015 - 2016
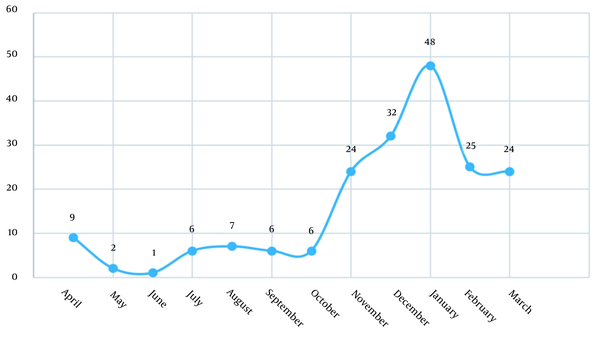
Categorizing samples based on the season showed that 13 (6.6) cases occurred in spring, 21 (10.7) in summer, 64 (32.7) in autumn, and 98 (50) in winter (Figure 1). In direct smear study, parasitemia rated from 1+ to 4+ and 9 (4.5) cases were 4+.
In molecular study, 61 clinical samples were positive for Leishmania spp. using PCR. In RFLP, 60 (98.37) sample patterns were identical to that of L. major and 1 (1.63) to that of L. tropica, compared to the reference strain (Table 3 and Figure 2). The cities with the highest rates of L. major infection were Shush (72.2), Dezful (19.6), and Gotvand (6.5), respectively. The only case of L. tropica was isolated from Dezful.
| Leishmania spp. | Dezful, No. (%) | Shush, No. (%) | Gotvand, No. (%) | Total, No. (%) |
|---|---|---|---|---|
| Leishmania major | 12 (19.6) | 44 (72.2) | 4 (6.5) | 60 (98.83) |
| Leishmania tropica | 1 (1.17) | 0 | 0 | 1 (1.17) |
| Total | 13 (20.77) | 44 (72.2) | 4 (6.5) | 61 (100) |
Distribution of Leishmania spp. Using PCR-RFLP Method Based on the Living Places of CL Cases in Dezful and Its Suburbs During 2015 - 2016
4.1. Phylogenetic Tree
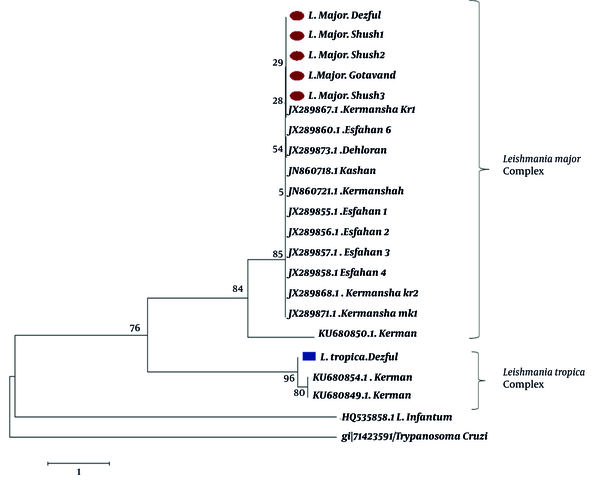
The phylogenetic tree from the sequences of ITS1 fragments clearly showed divergence between L. major and L. tropica isolates. Leishmania isolates were grouped into two main clades representing L. major and L. tropica. The single successfully sequenced L. tropica isolate from Dezful in the present study was placed in the cluster of other L. tropica sequences. Furthermore, homogeneity was observed among the L. major isolates from different geographical sites of our study. No clear grouping was demonstrated for the 20 isolates according to their geographical origins (Figure 3).
Bootstrap resampling values are shown at each branch. Isolates from the current study are marked by circles (L. major) and squares (L. tropica). L. infantum and T. cruzi DNA sequences were used as the out-group.
5. Discussion
Khuzestan is one of the most significant endemic areas of CL in southwestern Iran. Studies in this area have shown increased CL cases and genesis of new foci of the disease. In recent years, novel leishmanial foci, such as Dezful, Shush, and Gotvand, have been reported, indicating the potential spread of the disease in this area (24). In last decades, Dezful and its suburbs, as the second big city in Khozestan Province, was an endemic area of CL with low prevalence rates, but the disease rapidly increased from 372 new cases in 1994 to 1917 cases in 2015. Although studies have been carried out in different cities of Khuzestan, no information is available on the multiple circulating Leishmania spp. in the highlighted foci. Therefore, the current cross-sectional study was carried out during 2015 - 2016 on 196 Giemsa-stained slides from patients referred to health centers in Dezful, Gotvand, and Shush. Use of Giesma-stained slides is an appropriate method for field practices (22, 25). The method is advantageous in figuring out controversial cases or epidemiological studies that need integration.
Based on the current results, the number of infected males was greater than that of infected females. A similar study by Maraghi et al. (15) in Shush showed that the prevalence rate in males was higher than that in females. The greater prevalence in males might be a result of further outdoor working, less outfit covering, further traveling in desert areas, and greater possibility of contacting with sand flies (15). In 2014, Nilforoushzadeh et al. (26) showed that prevalence of the infection in males was two times greater than that in females, possibly due to the large number of males who worked as seasonal migrant labors or expatriates. Findings of the current study showed that the highest infected age group included 2 - 7 years old with a rate of 33.1%. This age group consists of children who usually play outdoor, and hence further expose to the causative agent. Maraghi et al. (15) in Shush showed that 42% of the CL infections occurred in children under 10 years old. Nejati et al. (25) reported a 41.6% rate for the infection, mostly in 15- to 24-year-old young people in Andimeshk. Nilforoushzadeh et al. (26) showed that the prevalence of leishmaniasis increased in individuals younger than 15 years old in endemic areas (26). The difference in results might be due to the restriction of the disease to native areas. Since under 10-year-old children are more likely to be infected with the pathogen in areas such as Shush, and hence acquire life-time immunity, infection in elder people is less possible (15).
In the current study, CL lesions were most frequently located on hands of the patients (50.5%), which was similar to that reported previously (15, 26). Studies from zoonotic CL areas in Iran and Saudi Arabia showed the maximum occurrence of lesions in uncovered body parts (15, 26). Findings from the present study showed that the seasonal pattern of the disease was in early winter (50%) between December to January (24.8%). Results from the previous studies showed that the highest and the lowest rates of seasonal leishmaniasis in Ahvaz, capital city of Khuzestan Province, were seen in autumn (49.2%) and spring (9%) respectively (27). Furthermore, results showed that most of the patients had only one lesion (60.3%) mostly on hands (50%). Results from other studies demonstrated that the number of lesions in leishmaniasis varied between 1 - 3 lesions per patient (28.3%) (28). Results from the molecular methods (ITS1-PCR RFLP) indicated that 98.4% of the CL cases in Dezful, Shush, and Gotvand were caused by L. major and 1.6% by L. tropica. Studies by Spotin et al. (29) in four areas of Khuzestan (North, South, West, and East) showed that 100% of CL cases were caused by L. major (30). Research by Maraghi et al. (30) demonstrated L. major as the predominant type of Leishmania spp. in Khuzestan. Maraghi et al. (15) reported that 90% of Leishmania spp. were L. major, which was similar to the reports from the present study. Other evidence including lesion type, number of ulcers, wet ulcer, seasonal distribution of the disease, and parasitism helped to verify these reports as most of the lesions were wet and existed in the lower parts of the body. These signs usually belong to rural form of leishmaniasis caused by L. major.
In general, prevalence of CL in Dezful, Shush, and Gotvand is remarkable. Despite low levels of genetic diversity in Iranian Leishmania isolates, sequence analysis was able to show at least one haplotype of L. major and L. tropica. No correlation was seen between the clinical symptoms in CL patients and genotype of the studied L. major isolates. In this study, the molecular methodology did not show any correlation between this haplotype and other Iranian L. major haplotypes from endemic areas. Indeed, results did not demonstrate any correlation between the haplotypes and the geographical areas, similar to the reports from other studies (31, 32).
5.1. Conclusions
Findings from this study show that L. major is the main agent of the re-emerged CL in Dezful and its suburbs and the disease is a zoonosis. Physicians and public health managers must be alerted, and hence further inter-sectorial collaborations are needed to limit primary reservoir hosts (gerbils particularly Tatera indica) in the studied areas. Moreover, further investigations and epidemiological studies on the disease are necessary.