1. Background
Diarrhea is caused by viruses broadly circulating in low-income countries and is the third leading cause of death in children younger than five years of age. Its highest incidence occurs in developing countries where there are 2 - 2.5 million deaths annually worldwide (1, 2). Several viruses including rotavirus (RV), norovirus (NoV), astrovirus (AstV), and adenovirus (AdV) account for a significant number of cases of acute gastroenteritis (3-6).
Recently, the association of enteroviruses with acute gastroenteritis has been reported (5, 6). Enteroviruses (EVs) are small, non-enveloped, single-stranded RNA viruses that belong to the Picornaviridae family (7, 8).
Enteroviruses are classified into poliovirus (PV) consisting of three serotypes, coxsackievirus type A (CV-A) with 23 serotypes, coxsackievirus type B (CV-B) with 6 serotypes, and echovirus (E) with 28 serotypes (7), besides some more recently discovered numbered serotypes. EVs show both endemic and epidemic patterns (8). Epidemics are typically caused by certain EV types such as E-30 (causing aseptic meningitis), EV-D70 (causing acute hemorrhagic conjunctivitis), EV-A71 (causing severe central nervous system), CV-A6 and CV-A10 (causing severe HFMD), and EV-D68 (causing flu-like symptoms) (8, 9). They are responsible for major public health concerns (9). EVs are considered a significant cause of many illnesses, including gastroenteritis, paralysis, meningitis, cardiomyopathy, respiratory tract infection, and the central nervous system infection (10-12). Although most infections are asymptomatic or cause less severe conditions, such as colds and fever, about 1.0% of EV infections result in severe diseases with high morbidity and mortality in young children and infants (10). EVs are transmitted through fecal-oral and respiratory routes.
The association of EV infection with diarrheal diseases is not well confirmed. However, the role of EV infection in acute diarrhea has been reported by a number of studies (5-10). Some EVs including E11, CVA6, CVB2, PV3, CVB4, E18, and CVA2 were detected in the stool of children with acute diarrhea in Hebei province, China (13). Moreover, the outbreak of gastroenteritis caused by echovirus type 6 was reported among children in Japan (14). E11 was similarly reported to be responsible for outbreaks of diarrhea in Malaysia (15) and southern India (16). Moreover, acute gastroenteritis caused by Coxsackie B-6 virus was confirmed in infants and children in Brasilia (17).
Another genus of picornaviridae named human parechovirus (HPeVs) has been found to be implicated in acute gastroenteritis (18). The role of EVs in acute diarrhea in children has not been yet investigated in Iran. However, the distribution of echovirus 30 and coxsackievirus A9 was formerly investigated among young neonates with sepsis in Ahvaz city, Iran (19). The complete sequence of echovirus 30 isolated from patients with meningitis has been detected and analyzed in Ahvaz (20). The high prevalence (59.09%) of EVs was also approved in children with aseptic meningitis in Ahvaz city, Iran (21).
2. Objectives
The detection of EVs is often not included in the pathogen surveillance of diarrhea, particularly in Iran. Nevertheless, based on the reported circulation of EVs in Ahvaz city, this study was conducted to evaluate the epidemiology and molecular characteristics of EVs circulating in outpatient children with acute diarrhea under < 5 years of age referring to Abuzar Hospital, Ahvaz, Iran.
3. Methods
At the initial stage of the study, about 300 liquidly fecal stool samples were collected from under-five children with acute diarrhea. All the patients were admitted to Abuzar Children Hospital, Ahvaz, Iran, during January - December 2013. Abuzar Hospital is the only referral children hospital in Ahvaz city. The inclusion criteria were samples negative for bacterial and parasitic pathogens, children younger than five years of age, and patients with acute diarrhea for less than 48 hours. The exclusion criteria were stool samples positive for bacterial and parasitic pathogens, the presence of WBC in the stool specimen, patients with mild diarrhea, and patients aged > 5 years. Based on the inclusion/exclusion criteria, 215 stool samples were excluded and the remaining 85 stool samples were kept at -70ºC for the prospective detection of EVs. Diarrhea is defined as ≥ 3 times/day defecation for less than 14 days (22). We collected data regarding the clinical characteristics of diarrhea, clinical history, physical examination results encompassing frequency and duration of diarrhea, vomiting, electrolyte disturbance, dehydration status, and the epidemiologic data including age, gender and treatment type. The data were recorded according to the WHO criteria (23).
All children presented clinical signs of gastroenteritis, i.e. fever and/or abdominal pain and/or diarrhea and/or vomiting. Diarrhea was the most frequently reported symptom (85/85: 100%), followed by vomiting (59/85: 69.41%), and fever (48/85: 56.47%). Abdominal pain was observed in 28 cases (32.94%).
3.1. Extraction of RNA and Preparation of cDNA
The viral RNA was extracted from the sample using a High-pure Viral Nucleic Acid Kit (Roche, Germany) following the manufacturer’s instructions. The cDNA was prepared (Fermentas, USA) according to the manufacturer’s instructions. The identification of the conserved 5’-UTR and/or the viral capsid sequence could differentiate between different EV species and types (24).
3.2. Semi-Nested PCR
Numerous EVs do not grow simply in cell culture. Hence, their diagnosis is based on PCR, which is more sensitive than traditional methods. In addition, PCR is more sensitive, reliable, and faster than culture. At present, PCR is the “gold standard” for EVs detection (25).
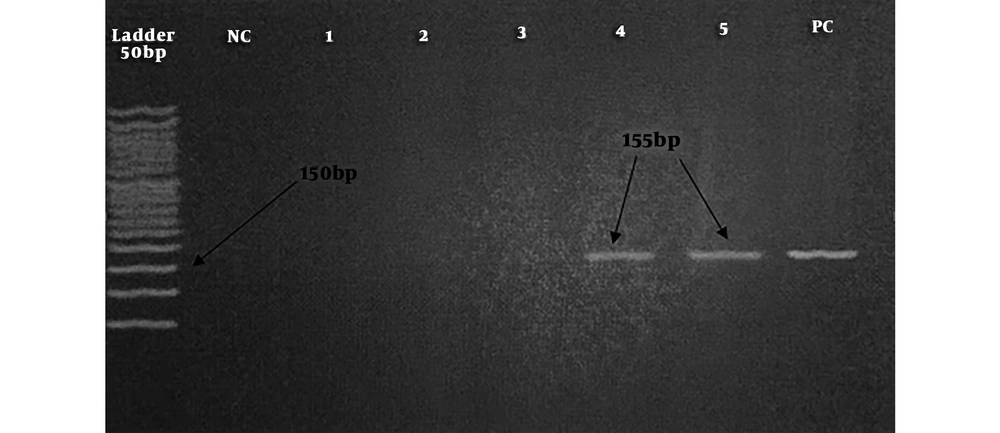
The semi-nested PCR was carried out for the detection of 5’-UTR region of EVs. The PCR reaction mixture contained 2.5 μL of PCR buffer, 0.5 μL of dNTP, 0.75 μL of each of EV-F1 and EV-R primers (Table 1), 0.75 μL of MgCl2, 0.12 μL of Taq polymerase, 2.5 μL of template, and up to 25 μL of distilled water (DW). The reaction mixture was transferred to a thermocycler (TC-512, Techne, UK) for 35 cycles of amplification (94°C for 45 seconds, 54°C for 45 seconds, 72°C for 45 seconds, and 72°C for 10 minutes). Then, 1 µL of the product was used as the template for the second round that contained 2.5 μL of PCR buffer, 0.5 μL of dNTP, 0.75 μL of each of EV-F2 and EV-R primers, 0.75 μL of MgCl2, 1 μL of template, 0.12 μL of Taq polymerase, and 18.62 μL of DW. The mixture was subjected to 35 cycles of amplification (94°C for 30 seconds, 58°C for 30 seconds, 72°C for 30 seconds, and 72°C for 5 minutes). The final product of 155 bp electrophoresed on the 2% gel indicated a positive result (21). The Sabin vaccine was used as a positive control.
3.3. Sequencing
To confirm the results of PCR and determine the EV serotypes, two positive PCR products were randomly selected and sequenced (Bioneer Company, South Korea). The sequences were blasted using available databases. A phylogenic tree was constructed with the neighbor-joining method using the partial nucleotide sequences of 5’-UTR region of EV positive samples. The reference sequences were retrieved from GenBank using their accession numbers.
3.4. Statistical Analysis
Statistical analysis was carried out for variables including gender, age groups, and season. The variables were recorded as means and standard deviation and analyzed by the chi-square test to determine the association of EVs with gender and season. Statistical analysis was performed using SPSS version 17.0 software. P values of < 0.05 were considered statistically significant.
4. Results
The patients’ age ranged between < 12 months and 60 months. The distribution of EVs among different age groups is presented in Table 2.
Overall, 21/85 (24.7%) children including 12/50 (24%) males and 9/35 (25.7%) females showed positive results for EVs (P = 0.3). The remaining 64/85 (75.29%) showed negative results for EVs. Figure 1 shows the results of the PCR assay.
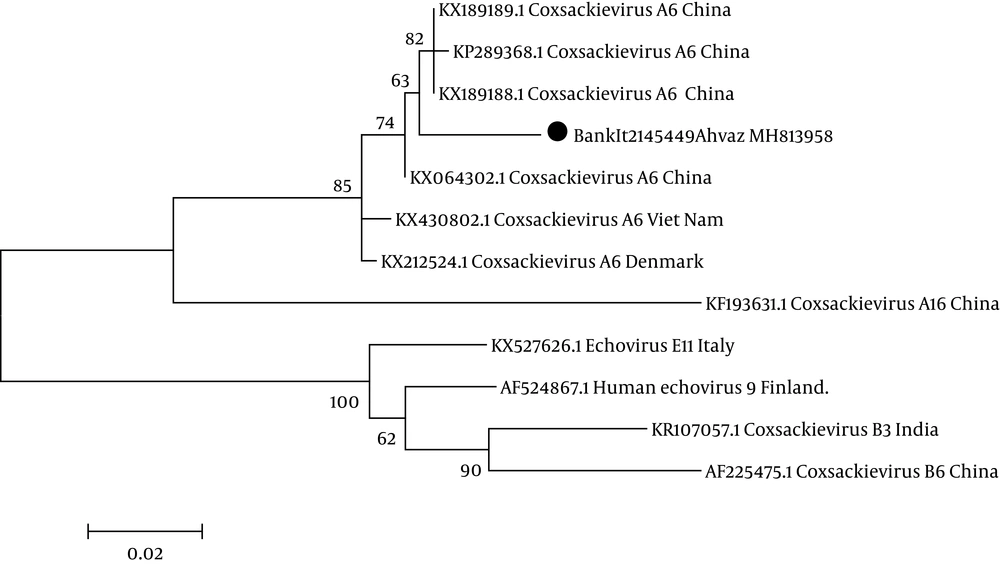
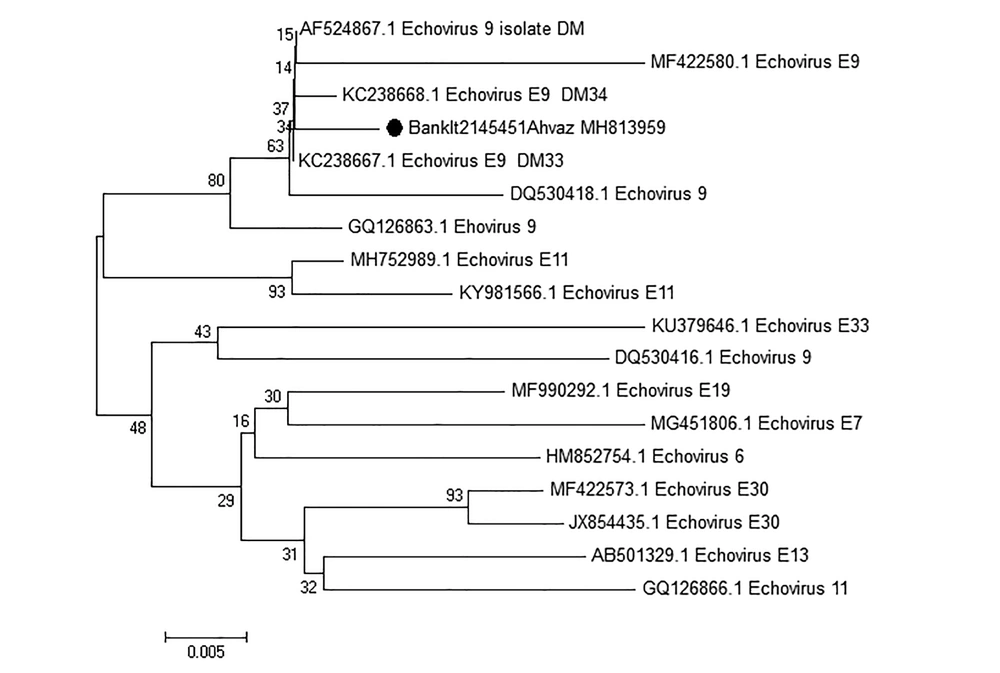
Based on the results of sequencing, one of the isolated samples from a two-year-old male was associated with coxsackievirus A6 with accession number BankIt2145449 Ahvaz MH813958 and the other one was echovirus 9 isolated from a one-year-old female patient with accession number BankIt2145451 Ahvaz MH813959. The results of the phylogenetic tree are demonstrated in Figures 2 and 3.
| Characteristics | EV Positive | Percentage | P Value |
|---|---|---|---|
| No. of patients (n = 85) | 21 | 24.7 | 0.3 |
| Female | 12/35 | 14.11 | |
| Male | 12/50 | 14.11 | |
| Age (months) | 0.468 | ||
| > 12 | 10/45 | 11.76 | |
| 13 - 24 | 3/16 | 3.52 | |
| 25 - 36 | 2/6 | 2.35 | |
| 37 - 48 | 0/3 | -- | |
| 49 - 60 | 6/15 | 7.05 | |
| Season | 0.001 | ||
| Spring | 0/10 | -- | |
| Summer | 0/11 | -- | |
| Autumn | 7/29 | 8.23 | |
| Winter | 17/35 | 20 |
The Profile of Patients with Acute Diarrhea Associated with EVs
Table 2 shows that the frequency of EVs was not found to be significantly different between males and females (P = 0.3). The distribution of EVs among different age groups was not significant (P = 0.468). The frequency of EVs proved to be the highest in winter (P = 0.001).
Neighbor-joining phylogenetic tree drawn with MEGA version 6 software based on the nucleotide sequences in the 5’-UTRregion of coxsackievirus A6 strain with the accession number BankIt2145449Ahvaz MH813958 labeled by black solid circle isolated from a patient with sepsis and comparison with other coxsackievirus A6 isolated from other regions of the world. Scale bar = 0.02.
Neighbor-joining phylogenetic tree of the nucleotide sequences in the 5’-UTRregion of Echovirus 9 strain with accession number BankIt2145451Ahvaz MH813959 isolated from a patient with sepsis in Ahwaz, Iran, labeled by the black square. It was compared with other echovirus 9 strains isolated from other regions of the world. Numbers in the branch are reproducibility after 1000 bootstraps. Scale bar = 0.005.
5. Discussion
The association between human enterovirus (EV) infection and acute gastroenteritis is well documented (5-18). In our study, 21/85 (24.7%) patients with acute diarrhea were positive for EVs. The prevalence of EVs in children with gastroenteritis has been reported in different parts of the world. Alcala et al. (Venezuela, 2018) detected EVs in 37.9% of children with gastroenteritis (5). Chitambar et al. (Mumbay, 2012) observed EVs in 40% of children with gastroenteritis, which was higher than the prevalence found in this study (26). Bubba et al. (Milan, Italy, 2015) found that 28/111 (25.2%) patients with gastroenteritis were EV-positive (27); this finding is in concordance with our results (24.7%). The frequency of EVs was not significantly different between males and females (14.11% vs. 10.58%; P > 0.05). In the present study, the obtained results of sequencing and blasting of the 5’-UTR region showed that one of the isolated serotypes from a two-year-old male showed 99% nucleotide identity to coxsackievirus A6 responsible for hand, foot, and mouth disease in China and Vietnam (28, 29), and 98% nucleotide identity to coxsackievirus A6 isolated in China (30). The role of coxsackievirus A6 in gastroenteritis in children was also reported by Chansaenroj et al. in Thailand (8). Another coxsackievirus A strain was also associated with gastroenteritis in children. Shobha et al. (Mumbay, 2012) reported coxsackievirus A13, 17, 19, and 21 in patients with gastroenteritis (26). The results of sequencing and blasting of the 5’-UTR region of the other EV strain isolated from a one-year-old female patient showed 96% nucleotide identity to echovirus 9 isolated from a small child at the clinical onset of type 1 diabetes in Finland and 94% nucleotide identity to echovirus 9 isolated from patients with aseptic meningitis in South Korea (31, 32). Other echovirus serotypes have been also associated with gastroenteritis in children. Shobha et al. (Mumbay, 2012) found echovirus serotypes 21 and 32 in patients with gastroenteritis (26).
The results of the phylogenetic tree exhibited that isolated coxsackievirus A6 was clustered with isolated coxsackievirus A6 causing hand, foot, and mouth disease in China (KX064302.1) and Vietnam (KX430802.1). The results of sequencing and blasting of the 5’-UTR region showed that isolated echovirus 9 showed 96% nucleotide identity to isolated echovirus 9 in Finland (KC238667.1).
In this study, EVs associated with acute diarrhea was detected in all age groups except for 37- 48-month age group. The highest rates of EVs were detected in patients’ age group of < 12 months (10/85; 11.76%) and 49 - 60 months (6/85; 7.05%).
The current study investigated the frequency of EVs associated with acute diarrhea and observed its occurrence only in autumn and winter seasons. The rate of EVs was found higher in winter than in autumn (P = 0.001). Shobha et al. (Mumbay, 2012) reported the high prevalence of EVs in autumn (40%) and winter 30% (26). Bubba et al. (Milan, Italy, 2015) documented the high prevalence of EVs in winter and autumn (27). To the best of our knowledge, it is the first report of EVs to cause acute diarrhea in Iran.
Viral intestinal infections are the most common cause of acute infectious diarrhea in children. Over the past decade, there have been major advances in the understanding of viral gastroenteritis etiology. Group A rotavirus is responsible for the majority of acute diarrhea cases in young children worldwide (28). Furthermore, other viruses like norovirus, adenovirus, enterovirus, bocavirus, sapovirus, astrovirus, calicivirus and more recently, torovirus and parechovirus have been associated with acute infectious diarrhea in children (33, 34).
5.1. Conclusions
In this study, a high prevalence of EVs (21/85; 24.7%) was detected in children with acute diarrhea, calling attention to the potential importance of EVs in diarrhea development. Based on sequencing results, one of the isolates in this study was identified as coxsackievirus A6 and the other isolate was echovirus 9. The remaining 64/85 (75.29%) showed negative results for EVs. Therefore, more studies are required to elucidate the distribution and characteristics of EV serotypes and their implications in this region. Based on the aforementioned results, it is suggested that the screening of enteroviruses be implemented by molecular methods such as PCR or real-time PCR in children with acute diarrhea.