1. Background
Bacteremia is the status, which is detected via a positive blood culture test with no contamination. It can be ignored if the condition is transient without clinical manifestation (1). However, bloodstream infection (BSI) with symptoms like septic shock and multiple organ dysfunction syndromes can be more problematic (2). In fact, bacteremia can cause morbidity and mortality, especially in patients with underlying diseases e.g., patients on hemodialysis (3). Although blood culture is the gold standard method to detect bacteremia, using previous antibiotics can lead to false-negative results. Different classes of antibiotics are often used to treat bacteremia, specifically in ICU; however, studies revealed that more than 30% of antimicrobial agents are unsuitable. Increasing the rate of antimicrobial resistance (AMR) could increase the rate of morbidity and mortality or at least the cost of treatment (4). Centers for Disease Control and Prevention (CDC) indicates that US$90 million in direct medical procedures and US$230 in total costs are considered additional expenses. Most of the costs are related to the long duration of treatment due to using inappropriate antibiotics and hospitalization (5). The average hospital stays in ICU and wards for patients with bacteremia are 2 - 7 days and 2 - 3 weeks, respectively. As mentioned, AMR is challenging in treating patients with bacteremia, especially in microorganisms like Staphylococcus aureus (S. aureus). Antibiotic resistance in S. aureus has become a difficulty in the healthcare system since 1940. Besides, methicillin-resistance S. aureus (MRSA) has been detected since 1960 (6). Besides, MRSA isolates also can transfer antibiotic resistance to other genera (7, 8). Another antibacterial resistance, which has been significant since 1987, is vancomycin-resistant Enterococcus (VRE). Treatment of the bacteremia caused by VRE is complicated because the side effects of some antimicrobial agents such as chloramphenicol are problematic. Moreover, the rate of resistance to antibiotics, including ampicillin and aminoglycoside is high (9). Therefore, controlling the transmission of VRE isolates in hospitals is crucial (6). Different bacteria may be detected in blood culture (10). The bacteremia due to Enterobacteriaceae is associated with increased mortality compared with BSI caused by Gram-positive species (11). Among Gram-negative bacteria, Acinetobacter and Pseudomonas spp. can result in severe nosocomial bacteremia (12). On the other hand, in some regions, including Iran, CoNS (coagulase-negative staphylococci) and S. aureus are the most frequent organisms isolated from blood culture (13). The AMR is a critical consideration for physicians to choose a suitable regimen. This is particularly important for the treatment of systemic infections because initial antimicrobial chemotherapy is almost empiric, and it must be based on knowledge of their antimicrobial susceptibility patterns. Early initiation of appropriate antimicrobial treatment is critical to decreasing morbidity and mortality among patients with BSI (14).
2. Objectives
The aim of this study was to investigate the rate and profiles of antimicrobial susceptibility of blood culture isolates from Tehran, Iran.
3. Methods
3.1. Patients and Sample Collection
In the current cross-sectional study, a total of 5,000 blood culture samples were collected from patients hospitalized in the Loghman General Hospital, Tehran, Iran, with positive blood culture results from 2012 to 2013. Blood samples were collected through cleaning of the venous site with 70% alcohol and povidone-iodine. The blood sample was injected into brain heart infusion and sodium thioglycollate broths in the ratio of one part of a blood sample to five parts of the culture broth (15).
3.2. Bacteria Isolation and Identification
The blood culture broths were sent to the laboratory and incubated at 37°C for seven days. MacConkey, blood, and chocolate agar media were subcultured at 24, 72 hours and on the 7th day. Then they were incubated at the appropriate temperature and atmospheres according to standard procedures (16). Isolated organisms were identified by conventional biochemical methods.
3.3. Antibiotic Susceptibility Test
The susceptibilities of blood samples against 15 antibiotics: trimethoprim-sulfamethoxazole (SXT: 1.25/23.75 µg), cefotaxime (CTX: 30 µg), cefoperazone (CP: 75 µg), ceftazidime (CAZ: 30 µg), imipenem (IMP: 10 µg), ampicillin (AMP: 10 µg), gentamicin (GM: 30 µg), chloramphenicol (C: 30 µg ), penicillin (P: 10 µg), oxacillin (OX: 1 µg), clindamycin (CC: 2 µg), erythromycin (E: 15 µg), vancomycin (V: 30 µg), ofloxacin (OF: 5 µg), and amikacin (AK: 30 µg) (Mast Group Ltd., UK) were tested by agar disk-diffusion method according to clinical and laboratory standards institute (CLSI) guidelines (17). E. coli ATCC 25922 was used as a control for the disk diffusion method.
3.4. Statistical Analysis
Statistical analyses were performed using SPSS software (version 25; SPSS, Inc., Chicago, IL, USA). The Fisher's exact test or the chi-square test was applied to analyze categorical data. A P-values < 0.05 in all experiments were considered statistically significant.
4. Results
4.1. Results of Sample Collection
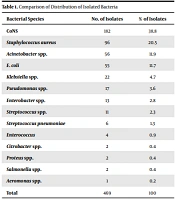
Of 5000 patients, 469 (9.3%) individuals had positive culture results. Also, 338 (72%) were male, and 131 (28%) were female. Of these 469 positive blood cultures, overall 469 isolates, 14 different species were detected. No mixed infection was observed. The most prevalent strains were CoNS (38.8%), S. aureus (20.5%), Acinetobacter (11.9%), and E. coli (11.7%). Pseudomonas, Enterobacter, Citrobacter, Proteus, Klebsiella, Salmonella, and Aeromonas spp. isolated in approximately 12% of cultures. Enterococcus and Streptococcus were found in 3% of positive cultures (Table 1). These results showed the predominance of bacteria, of which 63.75% and 36.24% were Gram-positive and Gram-negative bacteria, respectively.
| Bacterial Species | No. of Isolates | % of Isolates |
|---|---|---|
| CoNS | 182 | 38.8 |
| Staphylococcus aureus | 96 | 20.5 |
| Acinetobacter spp. | 56 | 11.9 |
| E. coli | 55 | 11.7 |
| Klebsiella spp. | 22 | 4.7 |
| Pseudomonas spp. | 17 | 3.6 |
| Enterobacter spp. | 13 | 2.8 |
| Streptococcus spp. | 11 | 2.3 |
| Streptococcus pneumoniae | 6 | 1.3 |
| Enterococcus | 4 | 0.9 |
| Citrobacter spp. | 2 | 0.4 |
| Proteus spp. | 2 | 0.4 |
| Salmonella spp. | 2 | 0.4 |
| Aeromonas spp. | 1 | 0.2 |
| Total | 469 | 100 |
4.2. Results of AMR Among Gram-Positive Bacteria
Sixty-two percent and 69% of CoNS were susceptible to gentamycin and fluoroquinolones, respectively, while 69% were resistant to cephalosporin. In addition, 88% of S. aureus isolates were MRSA, and 7% were VRE. Among Streptococcus spp., rather than enterococci and pneumococci, cefoperazone was active against 81% of isolates. Streptococcuspneumonia was highly susceptible to penicillin (75%) and resistant to trimethoprim-sulfamethoxazole (84%) (Table 2).
| Microorganism | NO. | V | IPM | CC | GM | C | E | CP | SXT | AK | AM | CAZ | CTX | OX | P | OF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CoNS | 182 | 89 | 46 | 43 | 62 | 36 | 2.5 | 69 | 32 | 1 | 9 | 0 | 31 | 2 | 1 | 1 |
| S. aureus | 96 | 86 | 63 | 60 | 62 | 39 | 49 | 69 | 62 | 2 | 4 | 1 | 50 | 12 | 0 | 0 |
| Streptococcus | 6 | 91 | 16 | 45 | 18 | 27 | 54 | 81 | 36 | 0 | 63 | 0 | 54 | 0 | 45 | 36 |
| S. pneumoniae | 22 | 83 | 66 | 33 | 0 | 66 | 16 | 83 | 16 | 16 | 50 | 0 | 66 | 0 | 75 | 100 |
| Enterococcus | 17 | 93 | 25 | 0 | 25 | 50 | 25 | 25 | 0 | 0 | 25 | 0 | 50 | 0 | 25 | 25 |
Abbreviations: SXT, trimethoprim-sulfamethoxazole; CTX, cefotaxime; CP, cefoperazone; CAZ, ceftazidime; IPM, imipenem; AK, amikacin; AM, ampicillin; GM, gentamicin; C, chloramphenicol; CC, clindamycin; E, erythromycin; V, vancomycin; P, penicillin; OX, oxacillin; OF, ofloxacin; CONS, coagulase-negative Staphylococcus aureus.
4.3. Results of AMR Among Gram-Negative Bacteria
As mentioned above, Acinetobacter spp. was the third common isolate. Moreover, 72 and 82% of isolates were resistant to imipenem, and amikacin, respectively, while 43% of Acinetobacter isolates were susceptible to cefoperazone. Further, 58 and 43% of E. coli were susceptible to amikacin, and gentamycin, respectively, and the rate of resistance to ceftazidime was 60%. The isolates of Klebsiella spp. were extremely resistant to more than three classes of antimicrobial agents, including cefotaxime (55%) (Table 3).
| Microorganism | NO. | V | IPM | CC | GM | C | E | CP | SXT | AK | AM | CAZ | CTX | OX | P | OF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter | 56 | 0 | 28 | 0 | 23 | 0 | 0 | 43 | 28 | 18 | 0 | 17 | 20 | 0 | 0 | 0 |
| E. coli | 55 | 0 | 62 | 0 | 43 | 0 | 0 | 54 | 32 | 58 | 1 | 40 | 29 | 0 | 0 | 0 |
| Klebsiella | 4 | 0 | 68 | 0 | 45 | 0 | 0 | 64 | 45 | 81 | 4 | 63 | 45 | 0 | 0 | 0 |
| Pseudomonas | 2 | 0 | 59 | 0 | 29 | 0 | 0 | 65 | 29 | 35 | 5 | 29 | 29 | 0 | 0 | 0 |
| Enterobacter | 11 | 0 | 0 | 0 | 84 | 7 | 0 | 69 | 77 | 69 | 0 | 54 | 69 | 0 | 0 | 0 |
| Citrobacter | 2 | 0 | 100 | 0 | 50 | 0 | 0 | 50 | 100 | 50 | 0 | 50 | 100 | 0 | 0 | 0 |
| Proteus | 13 | 0 | 50 | 0 | 100 | 0 | 0 | 50 | 50 | 50 | 0 | 100 | 50 | 0 | 0 | 0 |
| Salmonella | 2 | 0 | 100 | 0 | 100 | 50 | 0 | 100 | 50 | 100 | 0 | 100 | 50 | 0 | 0 | 0 |
| Aeromonas | 1 | 0 | 100 | 0 | 100 | 0 | 0 | 100 | 100 | 100 | 0 | 0 | 50 | 0 | 0 | 0 |
Abbreviations: SXT, trimethoprim-sulfamethoxazole; CTX, cefotaxime; CP, cefoperazone; CAZ, ceftazidime; IPM, imipenem; AK, amikacin; AM, ampicillin; GM, gentamicin; C, chloramphenicol; CC, clindamycin; E, erythromycin; V, vancomycin; P, penicillin; OX, oxacillin; OF, ofloxacin.
5. Discussion
The rate of bacteremia has been increased over the past years (1). Antimicrobial resistance (AMR) among bacteria isolated from blood culture is a worrying issue since it can influence the rate of mortality and morbidity (18). In Iran, AMR is a common cause of treatment failure of BSI. Since the determination of appropriate antibiotics is often not done in the best time, broad-spectrum antibiotics are used unnecessarily (19). Based on our results, Gram-positive organisms were the main pathogens (63.85%). Increasing rate of Gram-positive bacteria among blood cultures is seen in other regions (20). Bacteremia, which is caused by Gram-positive bacteria, is important because some microbial agents, including S. aureus, are often nosocomial and can make the situation of the patient more unfavorable. Besides, since some antimicrobial agents such as vancomycin has low tissue penetration, bacteremia caused by S. aureus often relapses (21). Increasing use of intravascular devices, as well as prolonged hospitalization, can result in raising the rate of the microorganisms, including CoNS and S. aureus (22). During the 1970s, BSI was most commonly associated with Gram-negative organisms, but recently, Gram-positive organisms began to emerge (23). This indicates that the organisms causing bacteremia are shifting toward Gram-positive in some regions (24). On the other hand, some studies reveal that Gram-negative bacteria remain predominant (25, 26). The critical issue is that Gram-negative bacteria can become problematic among patients with underlying diseases. Besides, previous use of antimicrobial agents in patients with underlying diseases can result in AMR among Gram-negative bacteria (27).
Besides the rates of MRSA, one of the most remarkable results in our study is high resistance to third-generation cephalosporins (3rd-GCs) among Gram-positive bacteria. Resistance to fluoroquinolone was almost variable among Gram-positive bacteria, which were 100% among S. aureus isolates and 0% among S. pneumonia. Furthermore, 3rd-GCs along with fluoroquinolones are generally used to treat BSI. As Lee et al. declared fluoroquinolones were more effective than 3rd-GCs for shortening the time of treatment of BSI (28); therefore, the high rate of susceptibility among S. pneumonia isolates can be considerable. Multi-drug resistance (MDR) is another issue that is important. All Gram-positive isolates were MDR, and specifically, the emergence of MDR among MRSA isolates can make the treatment more complicated and affect infection control in healthcare settings (29).
In addition, AMR was previously reported among Gram-negative bacteria (30, 31), which can be discussed from several points of view. Susceptibility to aminoglycoside was almost high among some Gram-negative isolates, including Salmonella, Aeromonas, Proteus, and Enterobacter. The effectiveness of aminoglycoside to treat the carbapenem-resistant isolates was studied in Shields et al. investigation (32). Some of the isolates showed high resistance to imipenem, including Enterobacter, which was susceptible to amikacin and gentamicin; therefore, it can give more chance to treat the patients more successfully. The rates of MDR isolates in Gram-negative bacteria are also impressive. Our Gram-negative isolates were totally resistant to ofloxacin, oxacillin, and clindamycin. The emergence of fluoroquinolone resistance along with beta-lactam drastically reduces the choice of treatment. Another important point is that previous use of antibiotics, including fluoroquinolone can influence increasing the rate of resistance in the future (33). Since the use of empirical antibiotics is a common way, previous use of antibiotics and subsequently the emergence of MDR can be threatening and challenging. The limitation of this study is the lack of molecular evaluation of antibiotics resistance-related genes, and it is suggested that minimal inhibitory concentration (MIC) and molecular tests should be performed for better assessment.
In conclusion, the results of current studies obviously indicate misuse of antibiotics in our society. National surveillance studies in Iran will be useful for clinicians to choose the right empirical treatment and will help control and prevent infections caused by resistant organisms.