1. Background
The characteristics of pluripotency can be achieved by ectopic expression of specific transcription factors in embryonic stem cell (ESC)-like cells can reprogram and generate induced pluripotent stem cells (iPSCs), which has created a new generation of therapy and research (1). The human iPSCs (hiPSC) and human ESCs (hESC) can be self-renewed and differentiated into all human cells when exposed to the proper developmental cues (2). Thus, these differentiated cells can be used as a source in a wide range of experiments to test potential treatments and drugs for diseases. Despite the high qualities of ESCs to differentiate toward different types of cells, the utilization of human ESCs has an ethical concern because they are acquired from the inner cell mass of blastocysts (3).
Unlike hESCs, iPSCs are generated from wholly differentiated adult donor cells. They contribute the same hereditary data with the donor and are detected as own cells by the donor’s immune system, which provides the inspiring ability for therapies with patient-derived iPSCs and does not need to suppress the immune system (4). Stem-cells are a reproducible and almost limitless source of transplantable cells and tissues used in disease therapies. All these ongoing and supreme improvements in the cell transplantation field require methods to generate high-quality cells, techniques to suppress the immune system, or strategies to differentiate stem-cells into favorable functional cells (5).
As the main modulator, autophagy is an extremely-conserved procedure, which helps attain accurate morphology and function of cells via the regulation of protein modification (6). Two processes for ubiquitin-like conjugation are needed to produce the autophagosomes. Autophagy-related gene (ATG) 5 is essential for both of these pathways. Recent reports have shown high rates of autophagy activity during iPSC derivation and maintenance (6). Furthermore, autophagy-related proteins (ATG) can increase autophagy and inhibit endoplasmic reticulum stress (7).
2. Objectives
The current study aimed to examine the mRNA expression of the main autophagy marker gene (ATG5) in different differentiation levels of iPSC toward insulin-producing cells.
3. Methods
3.1. Cell Culture
The human iPSC line (R1-hiPSC1-Royan Institute, Tehran, Iran) was cultured on the layer of mitomycin-C inactivated mouse embryonic fibroblasts (MEF) and fed with an iPSC medium. The culture medium comprised Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F12, L-glutamine, non-essential amino acids, β-mercaptoethanol, streptomycin, penicillin, ITS, b-FGF, and knockout serum replacement (KSR). The hanging drop method was conducted to generate embryoid body (EB) in the first step of differentiation. The iPSCs were differentiated toward definitive endoderm according to the method described with details in our previous study (8). The ATG5 gene expression was analyzed during differentiation, including iPSC, EB, mesendoderm (ME), and definitive endoderm (DE) stages.
3.2. Quantitative Real-time-PCR
Allprep RNA extraction kit (Qiagen, USA) and Takara (Japan) cDNA Synthesis Kit were used for total RNA extraction and complementary DNA synthesis. Quantitative RT-PCR (qRT-PCR) was performed via the StepOnePlus Real-time PCR system. We evaluated the relative mRNA expression levels by the 2-ΔΔCt method (ΔCt illustrates the difference between the ATG5 target gene and GAPDH control gene) and compared the fold changes with the non-treated hiPSC line.
The reaction mixture was made of CYBR green I master mix 2X (5 μL -Takara, Japan), ROX dye (0.2 μL), primers (10 pM each one), and 1 µL cDNA. Thermal conditioning was set as follows: one step at 95˚C (initial denaturation) for 30 s, then 40 cycles, including 95˚C for 5 s, 58˚C for 15 s, and 72˚C for 20 s. The sequence of forward and reverse primers of the ATG5 gene were 5’-GGCCATCAATCGGAAACTC-3’, 5’-AGGTCTTTCAGTCGTTGTCT-3’, and for GAPDH were 5’-GGACTCATGACCACAGTCCA-3’ and 5’-CCAGTAGAGGCAGGGATGAT-3’.
3.3. Statistical Analysis
The fold change calculation, descriptive statistics, and Graph design were performed by GenEX v.6.1, EXCEL v.2017, and GraphPad Prism v7.01 software, respectively. The different stages of differentiation were compared using One-way ANOVA with Tukey-Kramer’s post-hoc test. P-value less than 0.05 is considered significant.
4. Results
Our findings showed that the R1-hiPSC was well-cultured on the MEF layer and had a proper phenotype. The ATG5 gene expression level in several stages of cell differentiation compared with undifferentiated hiPSC. As described in previous reports, we examined the expression of pluripotency and endoderm markers to confirm cell differentiation. Our findings demonstrated that the cells were successfully induced to differentiate into endoderm.
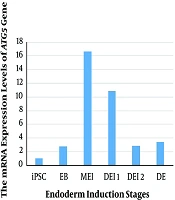
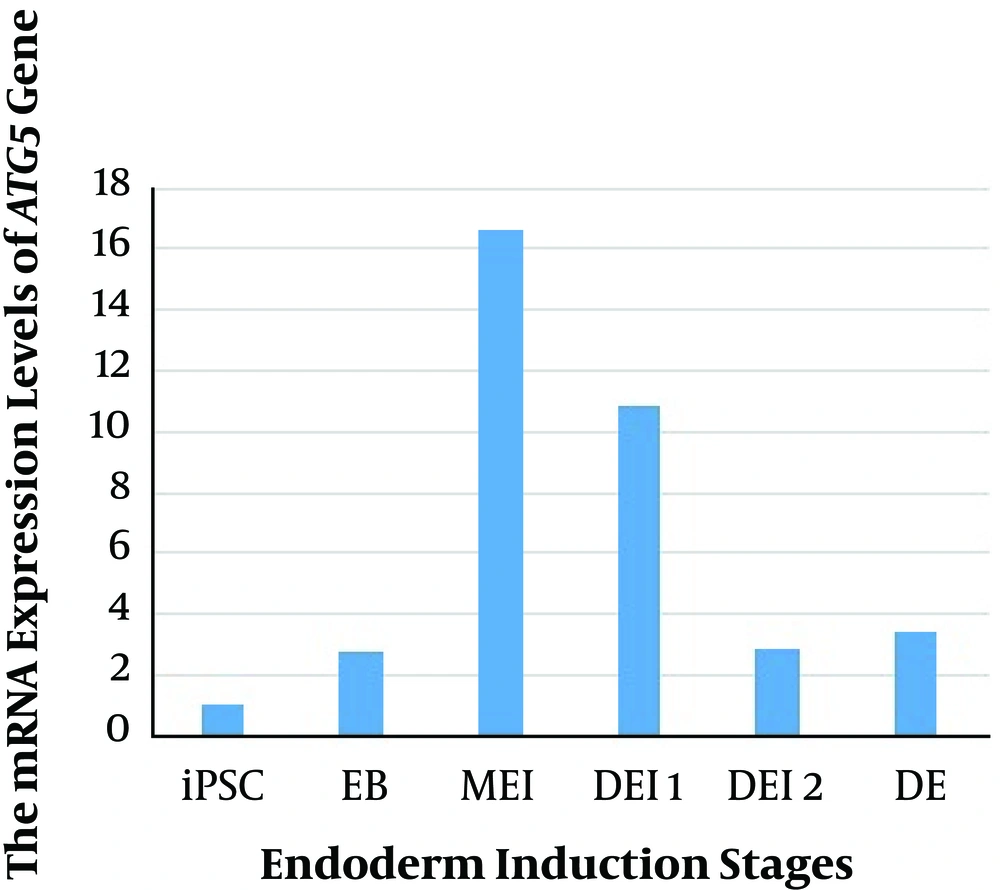
The mRNA expression of the ATG5 gene was examined in iPSC, EB, mesendoderm (ME), and definitive endoderm (DE) stages of differentiation (Figure 1). Our results showed the low levels of ATG5 mRNA expression in iPS cells. The maximum expression of the ATG5 was observed in the MEI stage (compared with iPSC and other stages of differentiation), and then the downregulated mRNA expression level was detected. However, the high level of the ATG5 expression was found in all stages of differentiation rather than iPSC. Therefore, the comparison of the ATG5 mRNA expression during differentiation indicated high levels of autophagy in the MEI stage.
5. Discussion
Autophagy is a significant, self-degradative procedure for the regulation of energy sources during development and stress status (9). Autophagy also eliminates misfolded or aggregated components and pathogens and clears damaged organelles (10). This procedure acts at a basal level in most tissues to maintain homeostasis. Moreover, autophagy is known as a quality control mechanism for proteins and organelles (11). It modulates several necessary cellular processes, such as self-renewal, differentiation, senescence, and apoptosis (12, 13). The role of autophagy in the maintenance and differentiation processes of stem cells as well as the generation of induced pluripotent have been previously shown (14).
Recent studies have demonstrated the conclusive role of autophagy in the function and survival of pancreatic beta cells (15). In vivo experiments showed that lack of autophagy in mice caused beta-cell mass reduction and decreased insulin secretion, demonstrating the role of autophagy in normal cell homeostasis (16, 17). Also, in vitro and in vivo studies showed that the over-induction of autophagy could reduce the function of cells (18). Moreover, there are some published reports, implying the vital role of autophagy during in vitro differentiation of stem cells (19-21). The ATG5 protein is essential in the development of early lymphocyte cells, the late activation of lymphocytes, and further plasma cell differentiation. According to previous reports regarding the role of ATG5 in cell differentiation, the autophagy genes status is a crucial subject in a pluripotent stem cell during several stages of differentiation (22, 23). In the present study, we differentiated hiPSC and analyzed the expression of ATG5 autophagy marker gene during endoderm induction (iPSC, EB, MEI, DEI 1, DEI 2, and DE). Our findings showed that autophagy was activated at the definitive endoderm generation step and reached a basic level during the last steps of differentiation reported by Pantovic et al. (24).
ATG5 is one of the recognized standards to study autophagic activity (25) and a significant mediator of autophagosome formation (26). ATG5 and ATG7 are needed to elongate and maturate the autophagosome. Imperfect depletion of ATG proteins impaired autophagy, evident by downregulation of LC3-II expression (27). The physiological effect of autophagy in pancreatic beta cells was examined by Ebato et al. with the generation of the mice with Atg7 deficiency. Their results showed that mice with β-cell specific autophagy deficiency (Atg7f/f:RIP-Cre, Atg7-deficient mice) showed impaired glucose tolerance with abnormal β-cell morphology (28).
Masakazu Sugiyama et al. observed that ATG5 silencing in Liver stem/progenitor cells reduced active LC3 and enhanced p62, showing autophagy inhibition and increased hepatic differentiation in the stem/progenitor cells. In contrast, SQSTM1/p62 silencing impaired hepatic differentiation (29).
Overall, our results demonstrated a time-dependent autophagy-specific gene expression during endoderm induction. We observed that the expression of the ATG5 (autophagy gene marker) gene was in the highest levels during the MEI stage. Determining the expression pattern of autophagy genes could be used to alter autophagy procedures and attain more efficient differentiation induction tactics.
5.1. Conclusion
The current study described mRNA expression of ATG5 autophagy gene during different days of differentiation (iPSC, EB, MEI, DE1, DE2, and DE). The results showed the decreased mRNA expression of ATG5 in the early stages of differentiation (EB) and then increased at the MEI stage. According to the results, autophagy was involved during the differentiation of iPSC and early stages of differentiation rather than the later stages. Subsequent studies are needed to find the precise role of autophagy in the differentiation of iPSC and obtain more efficient tactics to differentiate human iPSC.