1. Context
Similar to messenger RNA (mRNA), by using RNA polymerase II, lncRNA is transcribed (1) with or without polyadenylation, alternative cleavage, polyadenylation, and splicing, leading to divergent isoforms from the same locus (1, 2). Researchers have paid attention to long non-coding RNAs (lncRNAs) perform important functions in biologic pathways, such as epigenetic regulation, transcription of genes, transport of proteins, cell division, development of organs, cell replication, and chromosome dynamics (3, 4). Studies have shown that lncRNAs' dysregulation is associated with the evolution of multiple neurological disorders (4, 5). Nonetheless, in biological processes or diseases, many lncRNAs have general features, including structure, gene expression control functions, and molecule mechanisms (4, 6). We have compiled recent findings related to the main lncRNAs associated with neurological disorders.
1.1. Characteristics of Long Non-coding RNAs (LncRNAs)
Studies have indicated that lncRNAs from the Gencode v7 catalogue are produced in the same way as protein-coding genes when evaluating lncRNAs, although they display a pronounced bias towards two exon transcripts and are predominantly located at relatively low levels in chromatin and nucleus (7-10). LncRNAs are protected by a secondary structure, and they are thought to be evolved from different evolutionary pathways (11). They may be in positions other than the common shared elements, and just a few (~ 15%) lncRNAs display substantial sequence resemblance to other lncRNAs or protein-coding genes. Novel lncRNAs are derived from non-exonic de novo sequences or transposable elements rather than duplication (12). They have certain similar and common characteristics (1) lncRNAs transcribed with RNA polymerase II; (2) are loosely maintained at the sequence level and have a comparatively low level of expression and a pattern that is far more unique to cell tissue; (3) lncRNAs are controlled by transcriptional regulators (13).
1.2. Functions of LncRNAs
LncRNAs are well known for their involvement in the regulation of genes and genomic activity at different levels, including transcriptional, post-transcriptional, and translational. LncRNAs regulate the activity of genes through the cis mechanism that controls the activity of nearby genes and the ƒ mechanism that controls the activity of distant genes across the genome (14). In the latter case, for instance, telomeric repeat-containing RNA (TERRA) molecules arising from chromosome ends can regulate innate immune genes such as ISG15 in vivo through the impact of telomere location over long distance (TPE-OLD) (15). Inside the nucleus, multiple lncRNAs are situated where they can perform their critical functions through different mechanisms such as scaffolds and signaling (16).
2. Evidence Acquisition
This evaluation was compiled from different English papers retrieved from Elsevier, PubMed, and Google Scholar databases. The papers were searched using the following keywords: “brain disorders”, “ASDs”, “neurodegenerative diseases”, “nervous system”, and “lncRNAs”. Additional papers were recognized through screening the reference lists of relevant papers.
3. Results
The accumulated data shows that lncRNAs are involved in brain disorders. They may be used as molecular symptoms for cancer detection and drug projects for the treatment of nervous system diseases.
3.1. LncRNA General Functions in Brain
It has been shown that about 40% of lncRNAs are expressed directly in the brain (4000 - 20,000 lncRNAs). These brain-specific lncRNAs display the greatest signs of conservation of evolution compared to other tissues, which is a hallmark of functionality (7). Studies have shown that brain-expressed lncRNAs are enriched in predicted, maintained RNA structures and are more likely to have preserved functions (17). LncRNAs are shown to be expressed in a more cell-type particular way compare to protein-coding genes.
In general, 806 of 5195 lncRNAs have differential expressions across neuronal forms in transcriptome experiments on cortical pyramidal neurons, indicating their role in cell identity specification and conservation (18). Transcriptome analyses have shown that specific lncRNAs are expressed differently in the course of time and/or during development and adulthood in brain areas such as the hippocamp and cortex (19). Their preferable proximity to brain protein-encoding genes that are involved in transcriptional regulation or nervous system development is another trait of brain-specific lncRNAs (17).
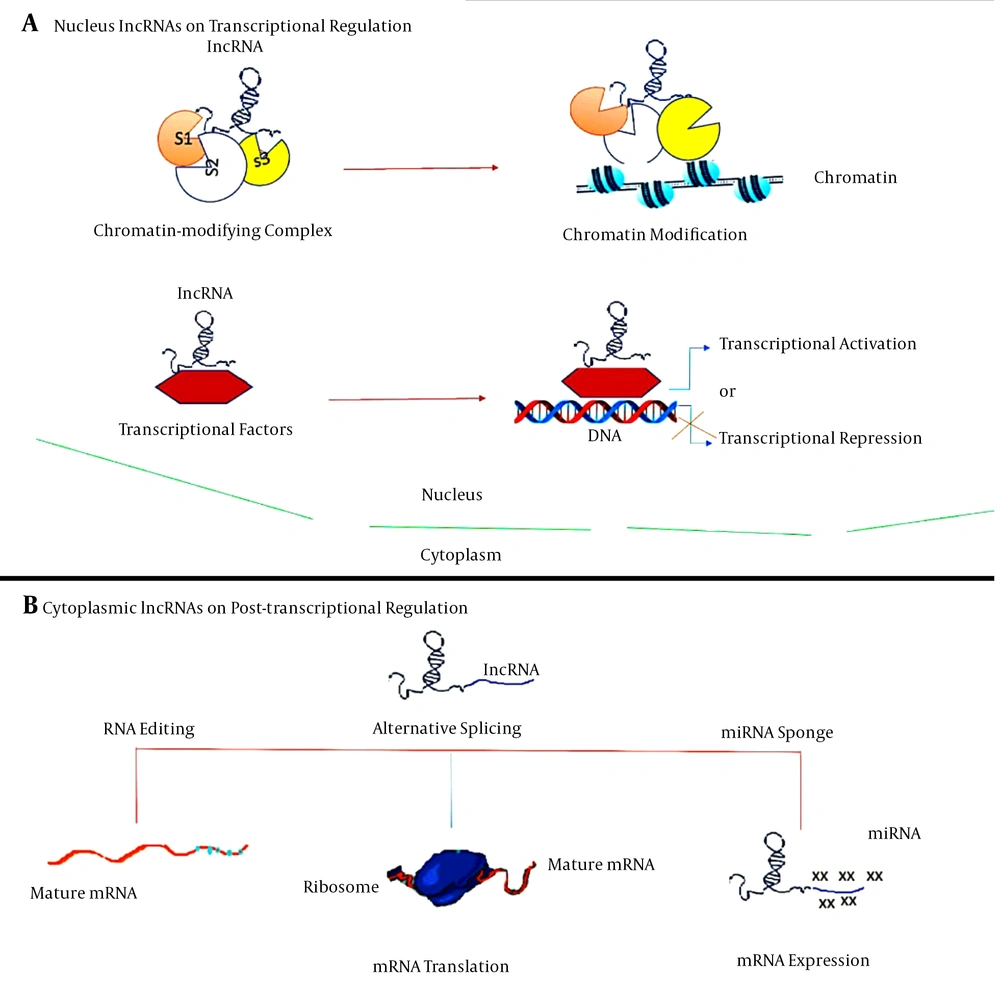
Given the nature and complexity of the central nervous system (CNS) of mammals, the brain can be used as the biggest source of lncRNAs relative to other somatic tissues. The specificity of these lncRNAs in tissues and cells contributes to the fate of the cell, ancestry determination, and preservation of the cells during the mammalian brain formation. The bulk of lncRNAs are found in the nucleus and can perform their regulatory roles through communicating with chromatin-modifying complexes as scaffolds for chromatin modifiers or by binding to transcription factors as transcriptional co-regulators (Figure 1) (20).
LncRNAs have been best identified at different stages for their role in controlling gene and genome function. LncRNAs theory mechanisms for gene and genome activity regulation. A, lncRNAs in the nucleus operate in transcriptional regulation by interacting with chromatin-modifying complexes or transcription factors; B, cytoplasmic lncRNAs usually function as RNA processing regulators, such as RNA editing, alternative splicing and miRNA-mediated mRNA expression.
3.1.1. Molecular Mechanisms of LncRNAs in Developing Central Nervous System (CNS)
Animal studies have demonstrated many of the lncRNAs’ functions, some of which are related to the CNS. LncRNAs, from early neural development to end-stage synaptic activity, play a crucial role in CNS development (21). It is known that balance between excitation and inhibition in the brain’s neural circuits is crucial to have a healthy condition. A well-known lncRNA is MALAT1 that is committed to regulating neurite growth. MALAT1 is found in several tissues; however, it is concentrated in nerve cells. MALAT1 can successfully recruit SR-family splicing proteins at transcription sites to start regulating synaptogenesis associated gene expression. MALAT1s knockdown results in reduced synaptic density (22).
3.1.2. LncRNA-Neuronal Disorders
More than 19000 lncRNAs play critical roles in multiple complex human diseases (23). The dysregulation of these lncRNAs has been shown to be correlated with the development of several different disorders such as Alzheimer’s disease (AD) (24). LncRNAs contribute to the pathogenesis of these diseases in various ways, from controlling transcription to modulating and translating RNAs.
3.1.3. Alzheimer’s Disease Pathology
Alzheimer’s disease is one of the most prevalent neurodegenerative disorders linked to aging that is characterized by the extracellular and intracellular tau protein and amyloid beta (Aβ) aggregation, respectively, as well as loss of synapses and neurons (25). Studies have confirmed the regulatory effect of lncRNAs in AD. For instance, a research showed that brain’s expression of lncRNAs is downregulated during AD (26).
3.1.4. BACE1-AS LncRNA
The antisense transcript of BACE1-AS, a conserved non-coding RNA derived from the reverse direction of the BACE1 locus on chromosome 11, has been strongly correlated with AD severity in both humans and transgenic mice (24, 27, 28). BACE1-AS controls BACE1 expression positively (29). Modarresi et al. showed that by knocking down BACE1-AS, the Aβ production and plaque deposition decreased, while neurogenesis markers increased. They concluded that this process occurs very early in amyloid pathogenesis and can serve as a therapeutic target/ or as an early biomarker.
According to the results of the study by Li, in APPsw transgenic cells, BACE1-AS mediates BACE1 expression and Aβ production upon addition of cell stressors, such as Aβ1-42. This result suggests that BACE1-AS can play a critical role in the progression of AD. In an attempt to determine whether BACE1-AS can be used as a blood-based biomarker in AD patients, Fotouhi et al. exhibited that plasma can serve as a potential source for BACE1-AS extraction and could be used as a potent biomarker of AD (30). In a continuing attempt to recover BACE1-AS additional roles, a new study by Xia et al. exhibited that BACE1-AS knockdown by siRNA in SAMP8 mice causes an increase in the proliferation of primary hippocampal neurons, which in turn, leads to improvements in memory and learning behaviors. They also found that following the depletion of BACE1-AS, BACE1, and APP protein production and tau phosphorylation are reduced in the hippocampus (31).
3.1.5. NAT-Rad18
The possible functions of NAT-Rad18, a natural transcription of antisense targeting Rad18 (a protein for DNA repair encoding gene), in the DNA repair mechanism for AD have been investigated. NAT-Rad18 has a widespread distribution in the brain of adult rats, with the highest expression in the cerebellum and cortex (32, 33). After exposure to Aβ, NAT-Rad18 was expressed in particular brain regions.
3.2. Schizophrenia (SZ)
Schizophrenia (SZ) is one of the most severe psychiatric illnesses diagnosed between the ages of 18 and 25 years for men and 25 and 35 years for women (34). Individuals with SZ have an abnormal perception of the reality, which may lead to a combination of hallucinations and disordered thinking and behaviors that affect their everyday functions. Numerous experiments have shown that non-coding RNAs are dysregulated in the CNS of these patients. This finding opens up new avenues to understanding the underlying molecular mechanism of neurodevelopmental disorders such as SZ (35).
3.2.1. HOXA Transcript Antisense RNA 2 (HOXA-AS2)
HOXA-AS2 interacts with Zeste homolog 2 enhancers (EZH2). EZH2 assumes a crucial function in the epigenetic silencing of cyclooxygenase-235, an enzyme that has been shown to be over expressed in SZ due to immune reaction dysregulation. An increase in EZH2 rate in the anterior cingulate cortex of SZ patients was found in RNA-seq data analysis. Such a function could be exercised by intervention in the natural developing brain (36, 37).
3.3. Epilepsy-LncRNAs
One of the most prominent neurodegenerative disorders marked by irregular electrical activity in the nervous system is epilepsy, which influences about 1% of the world's population (38-40). The function of lncRNAs has been explored in experimental animal epilepsy models. Scholars have found hundreds of lnRNAs expressed differently when comparing nervous tissue regulation in mice. Of those lncRNAs that were expressed differently, 54 (for pilocarpine) and 14 (for kainic acid) were similar to protein-coding genes, and it seems to cause major gene expression changes; thus, the potential Cis impact of these lncRNAs is indicated (41). LncRNAs function in Cis, silencing, or strengthening proximal gene expression on the same chromosome (42). Evidence suggests lncRNA‐UCA1 suppressed hippocampal astrocyte activation and JAK/STAT/GLAST expression in the rat models of temporal lobe epilepsy and improved adverse epileptic reaction (43). In the hippocampus and cortex, lncRNAs were dysregulated in a pilocarpine mouse model. The potential clinical targets for chronic epilepsy epigenetic modulation may be dysregulated lncRNAs with co-dysregulated mRNAs (44). Another study showed that lncRNA NEAT1 affected epilepsy inflammatory response by suppressing miR-129-5p and by further controlling Notch's signaling pathway in the IL-1β-induced cell model (45).
3.3.1. Brain-derived Antisense RNA Neurotrophic Factor (BDNF-AS)
Brain-derived antisense RNA neurotrophic factor (BDNF-AS) is an lncRNA transcribed from the opposite neurotrophic factor originating from the brain. The hippocamp in animal studies of epilepsy was significantly upregulated by BDNF mRNA and protein (38, 46). In the case of epilepsy, an investigation demonstrated that BDNF expression was up-regulated when the human neocortex is removed. In contrast, BDNF-AS was down-regulated following neocortex taking out. Such evidence indicates that the relationship between mRNA an lncRNA might reflect a human brain ability to change and adapt new states, and a possible therapeutic strategy for epilepsy could be used to inhibit BDNF pathways (38, 47).
3.4. Autism Spectrum Disorders (ASDs) -LncRNAs
The category of common neurodevelopmental disorders marked by disrupted social interaction and speech and restricted or repetitive actions or interests is autism spectrum disorders (ASDs) (48, 49). ASDs, clinically and etiologically, are complex and heterogeneous.
The involvement of lncRNAs in neurogenesis and GABAergic transmission suggests their potential role in ASDs pathogenesis. The expression of three lncRNAs in ASD patients' peripheral blood, namely p21-associated ncRNA DNA damage-enabled (PANDA), nuclear paraspeckle assembly transcript 1 (NEAT1), and taurine-upregulated gene 1 (TUG1), was assessed and compared with that of healthy subjects to establish peripheral biomarkers or targeted therapies for ASDs. In cellular apoptosis, the selection justification for these lncRNAs was their regulatory function. In ASD patients, the essential up-regulation of NEAT1 and TUG1 was observed (50).
3.4.1. NEAT1 RNA
NEAT1 RNA is preserved completely parallel to the paraspeckles in nuclei in foci. Such subcellular organizations are active in the nuclear processing of mRNAs. NEAT1 RNA interacts with the proteins of paraspeckles and seems to serve an important function in controlling the quantity of paraspeckles in proteins. By increasing miR-497 expression in the retina, NEAT1 was shown to control BDNF expression (5). Peripheral BDNF level in ASD patients has been shown to be considerably greater, which suggests the role of BDNF in ASDs’ pathogenesis. Elevated levels of NEAT1 could be involved in ASD pathogenesis via miR497/BDNF pathways (50).
3.4.2. TUG1
TUG1 was among lncRNAs having differential spatiotemporal expression in brain regions. In adult's brain, the TUG1 gene was expressed by wide range of neocortex cells.
Researchers have assessed the upregulation of multiple nearby genes in the brain tissues of Tug1 knocked-out mice, which suggests this lncRNA's Cis-acting role in controlling the expression of genes. This lncRNA in the course of an ischemic stroke changes cell apoptosis, inflammation, and angiogenesis. TUG1 has inhibitory effects on miR-9, an evolutionary retained miRNA associated with animal behavioral deficits. The abnormal TUG1 expression in ASD patients could be involved in ASD pathogenesis by the alteration of the expression of miR-9, and subsequently, multiple mRNAs are affected by this miRNA.
3.5. PD-LncRNA
The second most prevalent neurodegenerative disease that affects about 1 - 2% of the population over 65 years of age is Parkinson’s disease (PD). Non-motor signs, such as mood disturbances and panic, are often associated with motor signs including tremor, rigidity, and bradykinesia in PD patients (51, 52).
3.5.1. Uchl1-AS
Uchl1-AS is a nuclear lncRNA that is transcribed from the opposite strand to mouse ubiquitin carboxy-terminal hydrolase L1 gene. Uchl1 is a neuron-restrained protein that acts as a de-ubiquitinating enzyme and stabilizer of monoubiquitin. Uchl1 gene mutations were found to be linked to familial PD. Parkinson’s disease has confirmed Uchl1 protein oxidative inactivation (53). Uchl1-AS improves Uchl1 protein synthesis at the post-transcription stage, which relies on combined two-domain operations in the 5' antisense area that gives the specificity of the sensory target gene embedded repetitive element SINEB2 (short interspersed, nuclear factor subclass B3), which affords a protein synthesis activation domain (54, 55). Uchl1-AS activity is controlled via signaling. Uchl1 mRNA is mostly located in the cytosol, although Uchl1-AS is common in the nucleus of the dopaminergic neuron. MTOR suppressor-rapamycin therapy results in the recruitment of Uchl1 protein from the nucleus to the cytoplasm by the combination of Uchl1-AS. For developing parkinson's disease, the relationship between Uchl1 and ncRNAmTOR could be critical (55, 56).
3.6. Brain Cancers-LncRNAs
Several studies have established cancer-related lncRNAs (57, 58). It has been shown that they are exact p53 targets, including PINT, TUG1, PANDA, and linc-p21 (59, 60).
3.6.1. ANRIL's LncRNA
ANRIL's LncRNA is active in the tumor of the melanoma neural system and interacts with PRC2 to epigenetically suppress the p15 tumor silencer (61).
3.6.2. LncRNA CRNDE
In gliomas a strong upregulation of the lncRNA colorectal neoplasia differentially expressed (CRNDE) (62) and neuronal differentiation in induced pluripotent stem cells (iPSCs) occurs (63). The lncRNA gene MEG3 is a brain-specific inhibitor that suppresses cell growth and induces apoptosis controlled by p53; however, it is not present in pituitary tumors (64, 65) and meningiomas (66).
3.7. Multiple Sclerosis (MS)-LncRNA
Multiple sclerosis is an intense inflammatory disease that results in neuron sheath demyelination in the CNS (67). LncRNAs are active in MS development and control the distinction between B cells and CD4+ T-helper cells. BDNF plays an essential function in neuronal security as a growth-promoting gene. In an MS patient, microglia, astrocytes, and T cells have been shown to produce BDNF.
3.7.1. There Is an Important Interaction Between BDNF-AS as an LncRNA and BDNF
BDNF-AS functioning as lncRNA has been shown to inhibit BDNF in cells; also, it has functions as a negative BDNF regulator.
3.7.2. Growth Arrest Specific 5 (GAS5) Is Known as an LncRNA Active in Suppressing Glucocorticoid Receptors in MS Patients
In the immune system, glucocorticoids perform a significant function that may be viewed as a possible therapeutic agent for autoimmune diseases (68).
4. Conclusions
LncRNAs play a significant role in the development of brain disorders. The proof of lncRNAs’ association with brain disorders is mostly attributed to variations in expression. Research in the field of lncRNA has shown that the assessment of lncRNA expression levels in most common brain disorders can be used as a diagnostic and prognostic biomarker.