1. Background
Currently, more than six million unique protein sequences are stored in public databases, and this number is growing rapidly (ncbi.nlm.nih.gov/RefSeq). In addition, despite numerous advances in the determination of structures, only the structure of 50,000 proteins has been laboratory and experimentally validated. This apparent difference between the identification of protein sequences and the determination of their structure has led researchers to turn to computer-aided data analysis (1-4). The determination of the function of all these proteins by laboratory methods can be extremely time-consuming, costly, and even impossible in some cases. Two basic strategies are widely used to predict the role of proteins, one is based on the similarity of sequences and the other one is based on the structural similarity of proteins (5-7).
Unknown-function proteins are listed in different databases under different headings, including hypothetical, putative, and unknown proteins, which have a more well-known sequence regarding the order of the mentioned names. Hypothetical proteins are proteins that have been predicted by nucleic acid sequencing methods, and no chemical experimental evidence has shown the existence of these proteins; nevertheless, the existence of unknown proteins has been proven in vitro, although their role and function are still unknown. In this study, an unknown protein was randomly selected from the Arabidopsis Information Resource (TAIR) website (arabidopsis.org), belonging to the plant Arabidopsis thaliana.
Arabidopsis thaliana is a flowering plant in the Brassica family that includes economically important genera of Brassica and mustard. Arabidopsis thaliana was the first plant whose genome was sequenced. Although this plant is not important in terms of economic value, it is an ideal plant for research due to its small size, short growth period, and small genome. This plant has five chromosomes in the haploid stage, which has 135 Mb with 32,000 genes encoding proteins (8-11). In this study, an unknown protein with an access number or ID or AT2G15110.1 ID was selected from the TAIR site. Then, attempts were made using different databases to identify the properties and functions of the mentioned protein.
2. Methods
Firstly, the unknown protein was selected from arabidopsis.org, and then its access number was extracted from various databases. Operations, such as identification of the general properties of the desired protein, blasting the amino acid sequence, exploration of the motifs and domains in its amino acid sequence, examining the second, including alpha-helix and beta-sheets, and third structure of the target protein, recognition of the ligands, assessment of proteins involved with the target protein, and the determination of the protein location in the cell, were carried out.
3. Results and Discussion
In the initial investigation in order to understand the general characteristics of this unknown protein, the data shown in Table 1 were obtained.
| Target Protein Sequence with Access Number AT2G15110.1 on the TAIR Website | Properties | External IDs | ||
|---|---|---|---|---|
| Calculation of Molecular Weight | Calculation of Isoelectric Ph | Length In Terms of Amino Acids | ||
| MPPRKVVREVFLIDGKFEKYKTSLSTSSRLLLLRGAHQ IPHQIPLISPEPTVCPENPPPGHPSEEDRFSLSLNDLL QLYAVKKGRTKGTFFLSPRKGFRVFDDFPDKDEQWR KSYFFFPVNDLTYGNKTGLFVSEWAARTDLGWESLT IDRIRASGRRIRSRTDLAVSSPPFCPRIIDKADMSLPSS RQTTNKASVSAGKKPETPTSGSGKTIKDPAGKDSEKR AADKKRKQPEETNPSPPRSSRPRHEEKGAKLKGIVKE APQNLVVLSSRESETCESERRNVPLPAPPMTFADTMR TLVPPGSAIAPFDEMKEVNKENYLRFARKLGKLILEF NSVFCSHEDQLFDKDVEIESFKRSEDENAKAVEKAN KVMNRMKAAELQVQKLEVNNIDLTAKLKAGKNAY LDAIEKETQARADLRTCKEKMKKMEEEQAEMIVAA RTDERRKVRAQFHDFSSKYGNFFKESEEVETLKVRV AEAKANRELLEEIEKGEIPDLSKELESVRADEEKFAR HAAEPKTPRPDPTELTSLLADTPSEVAAESIPPAEVAII DEGGSNKGSTSEAGIAAMFPVDVEKDSGKTE | 64421.5 | 8.19 | 583 aa | GenPept: 238479252; UniProtKB: F4IHH0-1 |
General Characteristics of the Unknown Protein Identified by AT2G15110.1 on Arabidopsis Information Resource Websitea
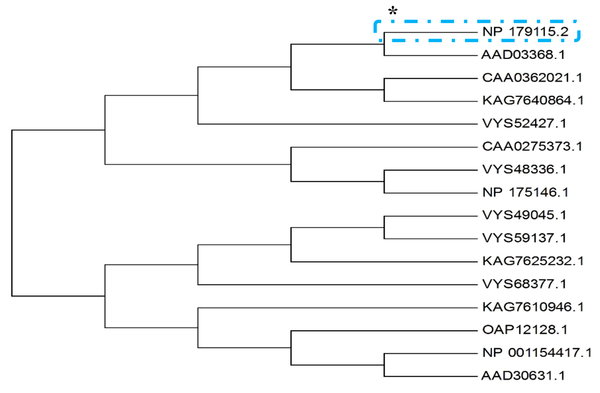
Protein blasting was then performed at the National Center for Biotechnology Information website (blast.ncbi.nlm.nih.gov). Among the records that had the most similarity with the target sequence, records 1 - 9 were unknown proteins. 15 of the records that were most similar to the target protein were selected and the phylogenetic tree of these records was drawn in MEGA 7 software (version 7) (12) after the alignment operation (Figure 1). It was observed that the unknown protein of interest did not has any common ancestor with any of the known proteins of these 15 records.
The unknown protein of interest blast was performed with dedicated access number of NP_179115.2 ** on the National Center for Biotechnology Information (NCBI) website. After the operations, the phylogenetic tree of these 15 records was drawn using MEGA software (version 7). It was observed that the protein in question had no common ancestor with any of the known proteins of these 15 records.
* The asterisk indicates the searched protein.
** This is the unknown protein of interest access number on the NCBI website that is different from that of the TAIR website.
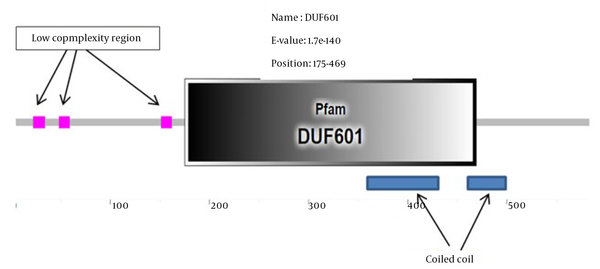
In the next step, the aforementioned protein was searched for motifs and domains in different databases. In all domain searched databases, it was observed that the searched protein had only a domain of unknown function (DUF) called DUF601 (Figure 2).
The DUF domains are protein domains that have no known function. This family of domains is collected in the Pfam database (pfam.xfam.org) and is prefixed with a DUF followed by a registration number, such as DUF2992 or DUF1220. The protein we are looking for has only one DUF and is named DUF601 in the Pfam database. This domain is located at position 186-469 of the amino acid sequence. Additionally, this unknown protein sequence had three low complexity region (LCR) (Figure 2). In proteins, LCRs are places where one or more amino acids are found in abundance. Due to their high abundance and potential for the ability to propagate in a short time through replication slippage, they can significantly contribute to increasing protein sequence length and producing new protein functions. However, little information is available on the overall impact of LCRs on protein evolution (13).
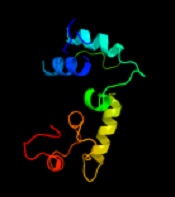
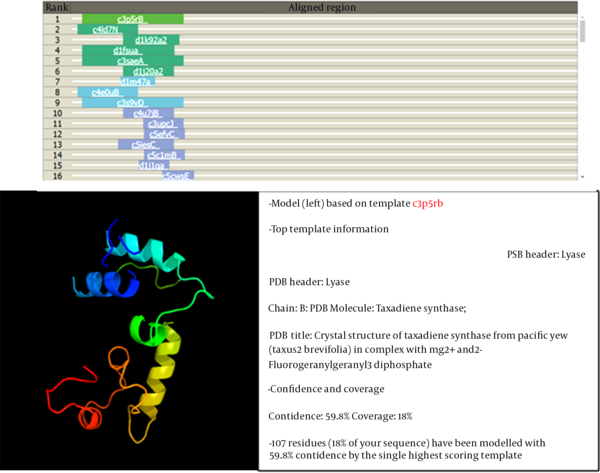
The identification of the function of the unknown protein was continued by examining its three-dimensional (3D) structure at the Phyre2 website (14), and the results are shown in Figure 3. At this stage, no significant similarity was observed between the desired protein and the proteins in the databases regarding 3D structure.
Among the identified models, the only model that was somewhat similar to the 3D structure based on the searched protein was c3p5Rb. This sample covered 18% of the searched protein, so its function could not be generalized to the searched protein.
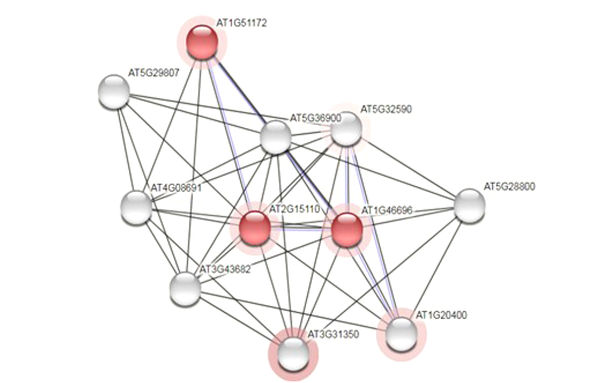
In the next step, the investigation was continued by following the relationship of the sought protein with other proteins that were carried out in UniProtKB database data (uniprot.org/uniprot) and from the STRING section. Figure 4 depicts the obtained results.
None of the proteins that interacted with the studied protein were known proteins, and only two of the mentioned proteins had the domain of unknown function (DUF601) in common with the target proteins, which are shown in red colour (Figure 4).
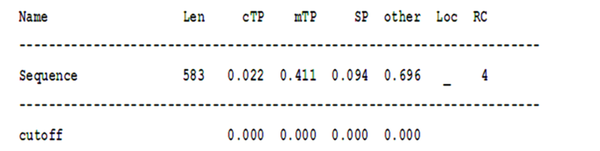
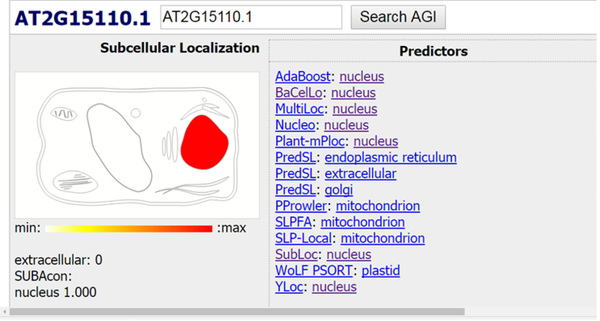
Finally, the location of the searched protein was examined using the data from different databases (15-17) in the cell, and various data were obtained. Most of the results confirmed the presence of the searched protein in the nucleus organ (Figures 5 and 6).
These data show that the presence of protein in chloroplast and mitochondrial organs and the secretion of that protein are very low, and the probability that it is in other organs is higher. This probability is 0.696; however, the validity of the results is low due to the value of RC, which is equal to 4.
Len, sequence length; cTP, chloroplast transit peptide; mTP, mitochondrial targeting peptide; SP, secretory pathway; a signal peptide; RC, reliability class, from 1 to 5, where 1 indicates the strongest prediction; Sign, meaning any other location
As it can be observed, most databases almost confirm the presence of the searched protein in the nuclear organ.
3.1. Conclusions
Based on the data obtained from various databases and the interpretation of these results, it was shown that the protein searched under access number AT2G15110.1 on the TAIR website bears no resemblance in sequence to known proteins and the 3D structure of proteins identified to date. Nevertheless, it turned out that this protein has only one DUF called DUF601, and there was the possibility of the presence of this protein in the nuclear organ due to its protein sequence in most databases of protein location identification.
However, to detect the function of this unknown protein, the researchers can repeat the same operations performed on the protein over time because countless new data are added to the databases every day, and this new data might help accurately identify the function of this protein. Since the requested protein has only one domain, clues can be provided to the function of this protein by knocking it out or knocking it down. DUF domains often lack basic functions and might not be recognizable by mentioned methods. Nevertheless, we can find its location and function by attaching reporter genes to it and transiently expressing it, or by using other methods to analyze protein function in a laboratory environment.