1. Background
Advanced cervical cancer (ACC) is regarded as the second most prevalent malignancy in women besides being the most common female-related cancer in dozens of developing countries (1). ACC patients are often treated with chemotherapy and radiotherapy followed by brachytherapy. Although chemotherapy and radiotherapy are invariably the main treatments for this type of tumor, the prognosis remains extremely poor (2). Radiotherapy is reported to be the cause of increased morbidity in ACC cases. Thus, this treatment is given only when the para-aortic nodal disease is confirmed (3). Hence, a precise evaluation of the extent of the tumor is substantial in order to design the most beneficial treatment. The majority of ACC cases contain human papillomavirus (HPV) infection, but viral infection was not detectable in a small percentage of the examined patients (4), reflecting the involvement of genetic variations as major risk factors for this disease (5) beside considering the sensitivity of ACC cells to genotoxic agents (6). Among ACC cell lines, Hela is the most widely used model cell line and no genomic reference for this cell line has been released so far. The analysis of the genomic content of Hela cells has uncovered a remarkably high level of aneuploidy. Moreover, gene expression patterns related to various cellular pathways (i.e., DNA repair pathways) has been reported to be altered in these cells (7). The cell death happens following the occurrence of DNA damage (8). Many DNA lesions are found to trigger the cell death including DNA double-strand breaks (DSBs) (9). The repair of DNA lesions is essential in preventing the cell death. Drugs that suppress DNA repair responses in cells harboring mutations in DNA repair pathway genes could be efficacious in monotherapy or in combination with other chemotherapeutic agents with the possible advantage of fewer side effects (10). DSBs are normally repaired by the homologous recombination (HR) pathway that involves RAD51 that requires breast cancer susceptibility proteins (BRCA1) and BRCA2 and poly (ADP-ribose) polymerase 1 (PARP1) (11). Studies revealed that the deficiencies of other HR-related proteins including CHK2, RAD superfamily, RPA1, ATR, ATM, and CHK1 sensitize the cells to PARP inhibition.
PARP1 has a critical role in more than one DNA repair processes, and small molecule selective inhibitors of PARP (PARPi) have been developed as chemotherapy sensitizers in cancer progression research (12). Olaparib (AZD2281, Lynparza) was the first PARPi approved by the food and drug administration (FDA) for the treatment of patients with deleterious germline BRCA-mutated advanced ovarian cancer (13). The growth-inhibitory efficacy of this PARPi was further investigated in other types of cancer cells as monotherapy or in combination with classic platinum anticancer drugs, which resulted in promising but limited outcomes due to the issues of drug resistance (14). In contrast, significantly lower levels of drug resistance were observed in treating HR-deficient breast cancer cells with AZD2461. In addition, anti-proliferative and apoptosis-inducing effects of AZD2461, as a newly developed structural analog for olaparib, have been recently discovered (15, 16). On the other hand, histone deacetylase inhibitors (HDACi) have been well established to exert anti-tumor effects on different cancerous human cells in vitro and in vivo (17). Valproic acid (VPA) is a conventional anti-seizure drug that recently has been demonstrated to have anticancer activities mediated by the selective inhibition of cellular histone deacetylase 1 (HDAC1) (18). VPA was reported to be able to suppress the cell proliferation as a single agent or in combination with other cell growth inhibitors, specifically in Hela cells (19, 20).
Combining conventional chemotherapeutic drugs with selective inhibitors of DNA repair pathway has recently gained much attention in the field of selective cancer treatment (21). While mutations in HR-related genes are associated with much higher risk of breast and ovarian cancers (22), these genetic variations are not frequent in other cancer types including ACC. The present experiment aimed to evaluate cell viability reducing and DNA repair potential of combined AZD2461 and VPA in Hela cells.
2. Methods
2.1. Chemicals and Assay Kits
3-(4,5- dimethylthiazol- 2-yl)-2,5-diphenyltetrazolium bromide (MTT), penicillin, streptomycin, amphotericin B, AZD2461, and VPA were obtained from Sigma-Aldrich (St. Louis, MO, USA). Both drugs were dissolved in HPLC grade DMSO and stored at -20°C as stock solutions for further use. Fetal bovine serum (FBS) was procured from Gibco (Rockville, MD, USA). HPLC grade DMSO, Trypan blue, RPMI-1640 medium, and trypsin-EDTA solution were purchased from INOCLON (G. Innovative Biotech Co. (INOCLON), Iran). Cell culture flasks were obtained from Biofill (Jet Biofill, China). All chemicals used were of analytical grade.
2.2. Cell lines and Culture Method
This study was conducted in spring 2018. Hela human advanced cervical cancer cell line was purchased from the Cell Repository of the Research Institute of Biotechnology, Ferdowsi University of Mashhad, Iran. The cells were cultured in RPMI-1640 culture media supplemented with penicillin (105 mg/mL), streptomycin (100 U/mL), Amphotericin B (2.5 mg/L), and 10% FBS. The cells were grown to confluence under stable culture conditions (a humidified atmosphere with 5% CO2 at 37°C incubator). The controls also were exposed to 1% DMSO and FBS containing fresh RPMI medium. All experiments were done at least in triplicate.
2.3. Evaluating Anti-Proliferative Effects of Each Inhibitor Using MTT Assay
In order to determine the half-maximal inhibitory concentration (IC50) of AZD2461 and VPA on Hela cells using the MTT assay protocol, 6000 cells/well were seeded and after one day of attachment period, the cells were treated with both inhibitors at increasing concentrations ranging from 0.612 mM to 20 mM and 6.25 μM to 200 μM for VPA and AZD2461, respectively. Following 24 and 48 hours of treatment, 5 mg/mL tetrazolium dye was added and the cells were incubated for two and a half hours. Then, the culture medium was removed, 180 μL of DMSO was added, and by using a microplate reader (Stat Fax 2100; Awareness Technology, Los Angeles, CA, USA), the absorbance at 492/630 nm wavelength was measured. The viability was expressed by dividing the absorbance of each concentration of treated cells by absorbance of control cells.
2.4. Analysis of Drug Combination
Hela cells were incubated with both inhibitors as single agents (VPA: 0.46 mM - 30 mM; AZD2461: 7.5 μM - 480 μM) with a combined ratio of 1:60, each diluted 1:2, within the given ranges. Using CompuSyn software (Version 1.0, Combo-Syn Inc., US), the interaction between constant ratios of both inhibitors was measured based on Chou-Talalay principles where regardless of the units of the drugs, CI = 0.9 - 1.1, CI < 0.9, and CI > 1.1 indicated an additive effect, a synergism, and an antagonism, respectively (23). The dose-effect relationships of combined inhibitors were modeled by using the median-effect equation:
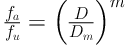
Where D indicates the dose of the inhibitor, m is the median effect coefficient, fa is the fraction affected, fu represents the fraction unaffected (fu = 1 - fa), and Dm is the median-effect dose (IC50 values) (24).
2.5. Morphological Examination of Hela Cells
Following 48 hours of incubation, Hela cells morphology and proliferation were monitored using an inverted phase-contrast microscope (Olympus CKX41, Tokyo, Japan) when treated with IC50 values of AZD2461 and VPA. Images were directly captured by a digital camera (Olympus C-7070).
2.6. Statistics
Results were analyzed using SPSS 22 software for Windows (release 22, SPSS Inc., Chicago, Illinois) and expressed as the mean ± standard deviation (SD). The values reported in the figures represent the means of an experiment repeated at least three times. P ≤ 0.05 was considered significant.
3. Results
3.1. Effects of AZD2461 and its combination with VPA on Hela Cells
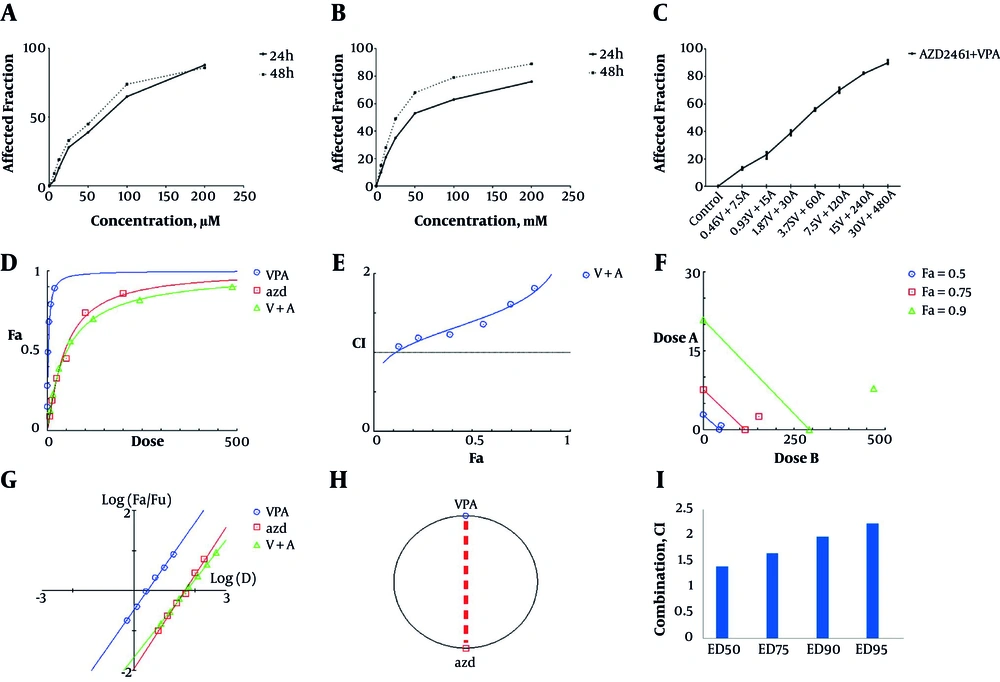
Both AZD2461 and VPA were able to decrease the viability of Hela cells in a time and concentration-dependent manner (Figure 1A and 1B) with IC50 values of 2.82 mM for VPA and 45.5 µM for AZD2461 following 48 hours treatment. Nevertheless, MTT result revealed that the combination of these inhibitors not only did not exert additive or synergistic effects, but also demonstrated mild antagonistic effects in previously described ranges (CI > 1.1). Dose-response plot, combination index plot, isobologram plot, median-effect plot, and polygonogram (at Fa = 0.5) for co-treatment with AZD2461 and VPA are depicted in Figure 2D - 2F, respectively. CI values of different effective doses (EDs) of both inhibitors were higher than 1.1 (Figure 2G). Consequently, co-treatment with AZD2461 and VPA was not found to be efficacious in reducing cell proliferation of Hela cervical cancer cells.
Concentration-response effects of AZD2461 and VPA on Hela cells using MTT assay following 48 hours treatment with both agents separately (A, B) and in combination (C) depicted by GraphPad Software. Dose-response plot (D), combination index plot (E), isobologram plot (F), median effect plot (G) and polygonogram (H) for co-treatment with AZD2461 and VPA are depicted by Compusyn software. Different effective doses (EDs) of VPA and AZD2461 combination indicate a mild antagonism (CI > 1.1) (G) as Hela cells were treated with various combinations of both inhibitors.
3.2. Analysis of Cell Morphology
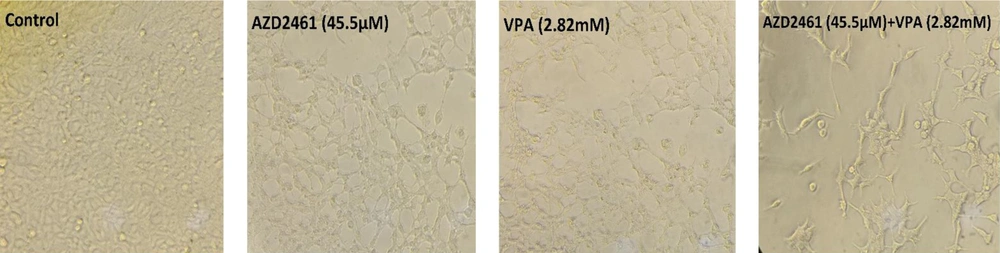
As shown in Figure 2, following 48 hours of treatment with AZD2461 (45.5 µM), VPA (2.82 mM), and AZD2461 + VPA (45.5 µM + 2.82 mM), the number of viable Hela cells apparently decreased compared to untreated cells, indicating that co-treatment with two inhibitors resulted in the increased rate of cell death. In addition, alterations in cell morphology such as shrinkage, rupture of cell membranes, and dropsy were evident in all three experimental groups.
4. Discussion
Genotoxic agents can block the replication fork during DNA replication process that leads to DSB formation (25). Hence, DNA damage response (DDR) of tumor cells against agents targeting DNA is expected to determine the efficiency of newly developed anti-cancer drugs. Alterations in DNA repair pathways could be the primary reason for special tumor cells to be dependent on limited DNA repair pathways to escape cell death. Moreover, mutations in DNA repair genes are proven to be associated with higher sensitivity of some tumor cells to certain DNA repair targeting agents such as PARP inhibitors (PARPi) since Poly (ADP-ribosyl) ation catalyzed by PARP1 is a prompt DNA-damage-dependent post-translational modification of nuclear proteins (i.e., histones HI, H2A, H2B, H3, and H4) that is essential for survival of damaged proliferating tumor cells (26). In 2010, Loser et al. discovered that null ATM-/-, Artemis-/-, and Ligase IV-/-p53-/-Hela cells display different responses compared to WT cells to low concentrations of two PARPi olaparib and KU55933, highlighting the importance of germline mutations of these genes in DNA repair pathway efficacy and clonogenic survival of Hela cells (27). In addition, a study conducted by Dejligbjerg et al. showed that VPA was able to exert anti-tumor effects in Hela cells due to its function as a class I‐specific HDAC inhibitor (28). Another experiment declared that VPA inhibits the proliferation of Hela cervical cancer cells through the activation of caspase-dependent pathways (19). Hence, no former study was done to assess the effects of the combination of PARPi and HDACi on cervical cancer cells while it seems essential to determine whether the effects of this combination are tumor specific and whether the therapeutic benefit can be predicted by the integrity of DNA repair pathways.
In contrast to an experiment conducted by Deben et al. (29), in which a combination of olaparib as a PARPi and APR-246 (PRIMA-1MET) synergistically induced cell death in P53-null lung cancer cells, our findings suggest a mild antagonistic interaction between VPA and AZD2461 Hela cells at all effective doses (CI > 1.1). Thus, this combination regimen was not able to effectively reduce Hela cells proliferation. Stankovic et al. reported that the combinatory effect of PARP inhibition and existing low-toxicity chromatin modifying agents (HDACi) could sensitize CLL tumors with DNA damage response defect (30). Konstantinopoulos et al. assessed the possible combination efficacy of olaparib and SAHA (Suberoylanilide hydroxamic acid) as a classic HDACi in ovarian cancer cells. It was concluded that SAHA could enhance olaparib anti-tumor activity by targeting homologous recombination DNA repair by decreasing Rad51 and BRCA1 expression. Combined with olaparib, SAHA induced apoptosis and pH2AX expression to a greater extent than either drug alone (31). Although many investigations reported that combining PARP inhibitors with other proliferation inhibiting agents could lead to better therapeutic results compared to monotherapeutic strategies (21), our findings call for further investigations regarding the evaluation of invasion-suppressing effects of the combination of VPA and AZD2461, using in vitro invasion assays. In addition, any alterations in the expression patterns of the genes involved in DNA repair or cell death pathways (i.e., caspase family, Beclin1) can be assessed using the real-time-PCR method. We will look forward to verifying our results by evaluating protein levels of some major apoptotic/necrotic genes besides confirming these data by flow-cytometric analysis or TUNEL assay for the detection of the type of cell death.