1. Context
Hepatocellular carcinoma (HCC), as the most common type of primary liver cancer, accounting for 70% - 90% of all liver cancers, is recognized as the seventh most common malignancy and the fourth cause of cancer-related death worldwide (1-4). Both environmental and individual genetic factors play important roles in the pathogenesis of HCC (5, 6). So far, several risk factors have been introduced for HCC, including chronic viral hepatitis (hepatitis B or C), non-alcoholic steatohepatitis (NASH), genetic predisposition with a history of HCC, and heavy alcohol or tobacco consumption (7, 8).
Previous research has confirmed the role of genes and metabolism-associated pathways in the development of HCC. Many upregulated and downregulated metabolic genes, such as those involved in the metabolism of carbohydrates and amino acids, are considered as key parameters in hepatocarcinogenesis (9). Glutathione S-transferases (GSTs), including glutathione S-transferase P1 (GSTP1), glutathione S-transferase T1 (GSTT1), and glutathione S-transferase M1 (GSTM1), are a specific group of enzymes, responsible for the detoxification of carcinogens (10).
Previous studies have shown that null deletions of GSTM1 gene may be a predisposing factor for lung, blood, and colorectal cancers (11, 12). Moreover, there is some evidence regarding the effects of GSTT1 null deletions and GSTP1 rs1695 polymorphism on the risk and prognosis of head and neck squamous cell carcinomas and lung cancer (13, 14). Identification of the role of GST family in the hepatocarcinogenesis can improve risk and prognosis prediction in patients with HCC. Besides, the development of novel gene therapies, including clustered regularly interspaced short palindromic repeats (CRISPR), highlights the importance of identifying the underlying genetic disorders.
Some original studies have investigated the association of GSTM1 null deletions, GSTP1 rs1695 polymorphism, and GSTT1 null deletions with HCC; however, they could not reach a definite conclusion, which might be due to the small sample size of these studies or limitations of methods and facilities. Therefore, in the present study, we aimed to assess the association of GSTM1 null deletions, GSTP1 rs1695 polymorphism, and GSTT1 null deletions with the risk of HCC.
2. Methods
2.1. Data Resources and Search Strategy
We systematically searched electronic databases, including PubMed, Scopus, and Web of Science, using combinations of the following keywords: “hepatocellular carcinoma”, “HCC”, “GSTM1”, “GSTT1”, and “GSTP1” (Appendix 1 in Supplementary File). First, a systematic search was conducted in November 2018, which was updated in March 2019. All selected articles were written in English. Besides, we screened the reference lists of all included studies for identifying any additional papers.
2.2. Eligibility Criteria
We included case-control studies that assessed the relationship between HCC and GSTM1 null deletions, GSTP1 rs1695 polymorphism, and GSTT1 null deletions. The diagnosis of HCC was mainly confirmed by imaging and measuring the serum alpha-fetoprotein level in all included studies. Studies that had not reported the exact number of patients with polymorphism genotypes in each group were not included. Also, studies without healthy controls were excluded. Healthy controls were defined as hospital- or community-based people with no history of liver diseases.
2.3. Study Selection, Quality Assessment, and Data Extraction
Two authors (M.H.K and H.S.) independently reviewed all identified papers (15). Any disagreements between the authors were resolved by neutral discussion. The Newcastle-Ottawa scale was applied for the evaluation of case-control studies (16). Next, the publication details and data about the patients were extracted from each study. Also, the first author’s name, publication year, sample size, age, gender, and underlying diseases of patients, and frequency of each polymorphism genotype in each group were determined.
2.4. Data Analysis
For determining the heterogeneity of data, chi‐square and I-square (I2 range: 0% - 100%) tests were performed. P-value less than 0.1 was considered statistically significant for the heterogeneity of data, based on the chi‐square test. If I2 was less than 50%, there was no significant heterogeneity, and the fixed‐effects model was used. On the other hand, if I2 was above 50%, the random‐effects model was selected to calculate the pooled odds ratio (OR), 95% confidence interval (CI), and P‐value. Statistical analysis and generation of forest and funnel plots were performed in Review Manager version 5.3.
3. Results
3.1. Study Screening
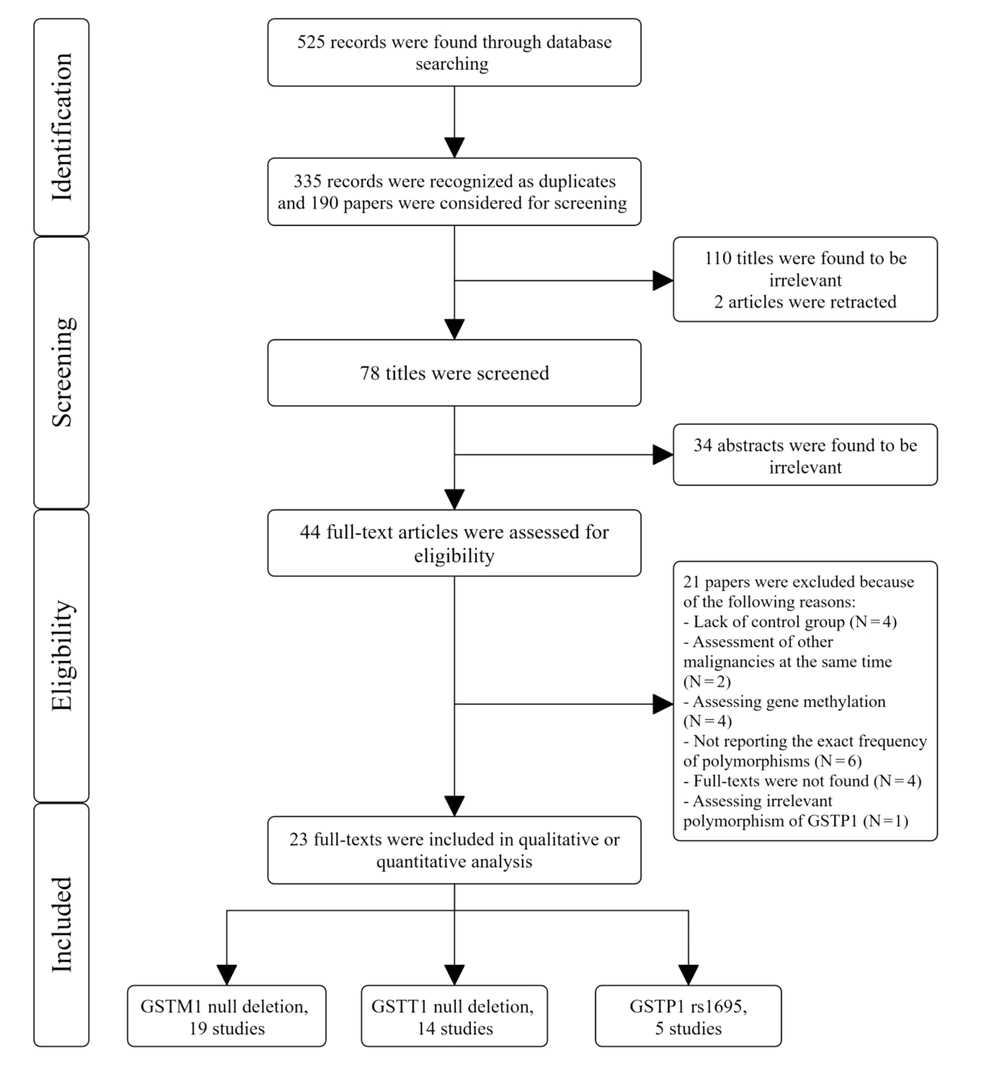
After searching the databases, we found 525 records and investigated 190 papers after removing duplicates. We excluded 112 articles by title screening and 34 articles by abstract screening. Also, during title and abstract screening, we excluded 37 articles owing to the study design (not case-control). Finally, after updating our search, 23 studies were included in this review (Figure 1).
3.2. Risk of Bias Assessment
The results of quality assessment for the case-control studies, based on the Newcastle-Ottawa scale, are shown in Table 1. All case-control studies were categorized as low risk. No study was excluded after this assessment.
| First Author | Selection | Comparability | Exposure |
|---|---|---|---|
| Abo-Hashem et al. (17) | **** | * | *** |
| Asim et al. (18) | *** | * | *** |
| Bian et al. (19) | *** | * | *** |
| Boccia et al. (20) | *** | * | *** |
| Borentain et al. (21) | **** | * | *** |
| Chen et al. (22) | **** | * | *** |
| Gelatti et al. (23) | *** | * | *** |
| Imaizumi et al. (24) | **** | * | *** |
| Kao et al. (25) | **** | * | *** |
| Kiran et al. (26) | **** | * | *** |
| Kirk et al. (27) | **** | * | *** |
| Ladero et al. (28) | **** | * | *** |
| Ladero et al. (29) | **** | * | *** |
| Li et al. (30) | **** | * | *** |
| Dai Long et al. (31) | **** | * | *** |
| McGlynn et al. (32) | **** | * | *** |
| Munaka et al. (33) | *** | * | *** |
| Sophonnithiprasert et al. (34) | **** | * | *** |
| Sun et al. (35) | **** | * | *** |
| Tiemersma et al. (36) | **** | * | *** |
| Wei et al. (37) | *** | * | *** |
| Yu et al. (38) | **** | * | *** |
| Ma et al. (39) | *** | * | *** |
Quality Assessment of Studies Based on the Newcastle-Ottawa Scale
3.3. Characteristics of the Selected Studies
The included papers were case-control studies published after 1995. These studies assessed the effects of all or at least one GSTM1 null deletion, GSTP1 rs1695 polymorphism, and/or GSTT1 null deletion on the risk of HCC. The characteristics of these studies are presented in Table 2.
| First Author (Reference) | Country | Ethnicity | Publication Year | HCC Cases, N | Control, N | Evaluated Polymorphism(s) | GSTM1 Genotype in HCC, null/Present, % | GSTM1 Genotype in Control, Null/Present, % | GSTT1 Genotype in HCC, Null/Present, % | GSTT1 Genotype in Control, Null/Present, % | GSTP1 Genotype in HCC, IV+VV/II, % | GSTP1 Genotype in Control, IV+VV/II, % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abo-Hashem et al. (17) | Egypt | Non-Asian | 2016 | 40 | 40 | GSTP1 | - | - | - | - | 42.5/57.5 | 22.5/77.5 |
| Asim et al. (18) | India | Non-Asian | 2010 | 254 | 525 | GSTM1 GSTT1 | 59.8/40.1 | 29.9/70.1 | 38.6/61.4 | 16.8/83.2 | - | - |
| Bian et al. (19) | China | Asian | 2000 | 63 | 88 | GSTM1 GSTT1 | 57.1/42.9 | 42/58 | 87.3/12.7 | 62.5/37.5 | - | - |
| Boccia et al. (20) | Italy | Non-Asian | 2015 | 221 | 290 | GSTM1 GSTT1 | 52.2/47.8 | 51.9/48.1 | 29.9/70.1 | 23.9/76.1 | - | - |
| Borentain et al. (21) | France | Non-Asian | 2007 | 56 | 89 | GSTM1 | 46/54 | 61/39 | - | - | - | - |
| Chen et al. (22) | Taiwan | Asian | 2010 | 177 | 386 | GSTP1 | - | - | - | - | 36.7/63.3 | 29.5/70.5 |
| Gelatti et al. (23) | Italy | Non-Asian | 2005 | 200 | 400 | GSTM1 GSTT1 | 49.5/50.5 | 53.8/46.3 | 16/84 | 18/82 | - | - |
| Imaizumi et al. (24) | Japan | Asian | 2009 | 209 | 256 | GSTM1 | 52.6/47.4 | 55.8/44.2 | - | - | - | - |
| Kao et al. (25) | Taiwan | Asian | 2010 | 102 | 386 | GSTM1 GSTT1 | 52.9/47.1 | 54.7/45.3 | 50/50 | 51.8/48.2 | - | - |
| Kiran et al. (26) | India | Non-Asian | 2008 | 63 | 169 | GSTM1 GSTT1 | 25.4/74.6 | 22.5/77.5 | 27/73 | 14.2/85.8 | - | - |
| Kirk et al. (27) | Gambia | Non-Asian | 2005 | 216 | 408 | GSTM1 GSTT1 | 29.5/70.5 | 25.6/74.2 | 47/53 | 44.9/55.1 | - | - |
| Ladero et al. (28) | Spain | Non-Asian | 2007 | 184 | 248 | GSTP1 | - | - | - | - | 52.2/47.8 | 51.4/48.6 |
| Ladero et al. (29) | Spain | Non-Asian | 2006 | 184 | 329 | GSTM1 GSTT1 | 47.8/52.2 | 45.3/54.7 | 28.8/71.2 | 23.1/76.9 | - | - |
| Li et al. (30) | China | Asian | 2012 | 476 | 481 | GSTM1GSTT1GSTP1 | 51.3/48.7 | 43.9/56.1 | 25.2/74.8 | 19.6/80.4 | 59.5/40.5 | 56.1/43.9 |
| Dai Long et al. (31) | China | Asian | 2006 | 257 | 649 | GSTM1GSTT1 | 69.65/30.35 | 48.07/51.93 | 56.81/43.19 | 45.76/54.24 | - | - |
| McGlynn et al. (32) | China | Asian | 1995 | 52 | 116 | GSTM1 | 56/44 | 41/59 | - | - | - | - |
| Munaka et al. (33) | Japan | Asian | 2003 | 78 | 138 | GSTM1GSTT1 GSTP1 | 38.5/61.5 | 49.3/50.7 | 51.3/48.7 | 47.8/52.2 | 23.1/76.9 | 33.3/66.7 |
| Sophonnithiprasert et al. (34) | Thailand | Asian | 2019 | 49 | 66 | GSTM1 GSTT1 | 69/31 | 61/39 | 24/76 | 38/62 | - | - |
| Sun et al. (35) | Taiwan | Asian | 2001 | 79 | 149 | GSTM1GSTT1 | 37.7/62.3 | 60.2/39.8 | 44.8/55.2 | 77/51 | - | - |
| Tiemersma et al. (36) | Sudan | Non-Asian | 2001 | 112 | 194 | GSTM1 GSTT1 | 42.7/57.3 | 38.8/61.2 | 35.8/64.2 | 37.8/62.2 | - | - |
| Wei et al. (37) | China | Asian | 2012 | 181 | 641 | GSTM1 GSTT1 | 65.2/34.8 | 47.6/52.4 | 57.5/42.5 | 43.1/56.9 | - | - |
| Yu et al. (38) | Taiwan | Asian | 1995 | 30 | 150 | GSTM1 | 53.3/46.7 | 63.3/36.7 | - | - | - | - |
| Ma et al. (39) | China | Asian | 2001 | 120 | 140 | GSTM1 | 59.1/40.9 | 46.4/53.6 | - | - | - | - |
The Characteristics of the Included Studies
3.4. Outcome Evaluation
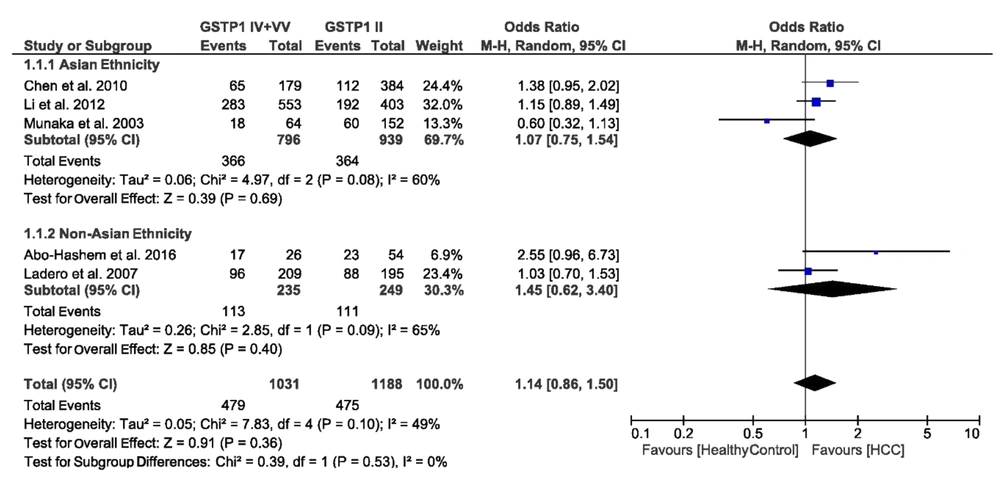
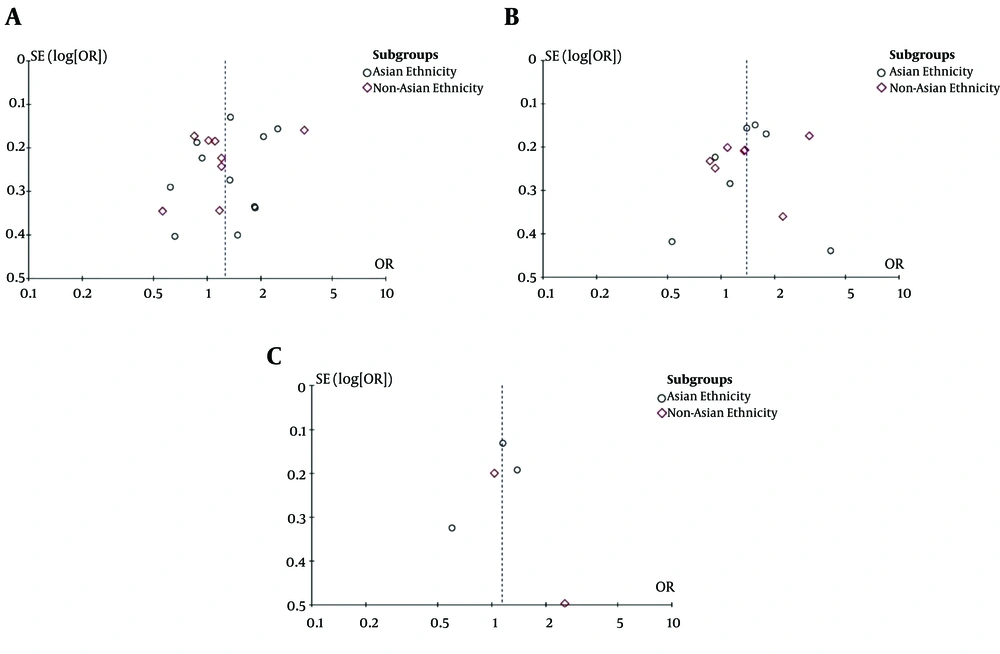
We evaluated the association of GSTM1 null deletions, GSTP1 rs1695 polymorphism, and GSTT1 null deletions with the risk of HCC. Figures 2-4 present a summary of the results for GSTM1 null deletions, GSTT1 null deletions, and GSTP1 rs1695 polymorphism as forest plots, respectively. Figure 5 shows the funnel plots for all of the assessed polymorphisms.
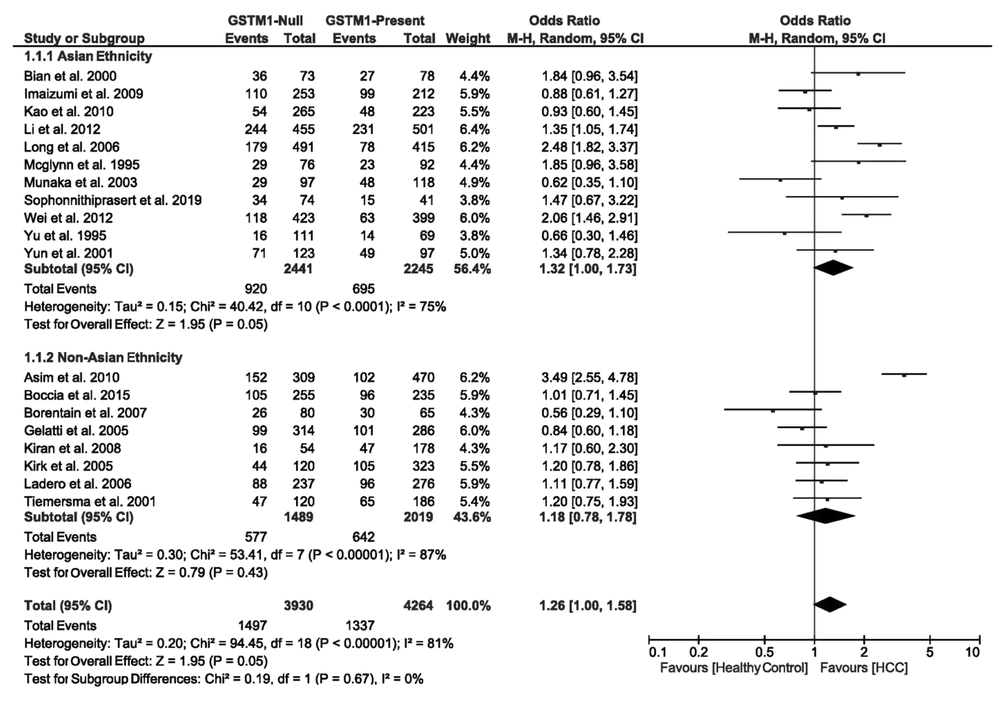
3.4.1. Association of GSTM1 Null Deletions with the Risk of HCC
We found the heterogeneity of data regarding the association of GSTM1 null deletion with the risk of HCC in both Asian (I2 = 75%, P < 0.0001) and non-Asian (I2 = 87%, P < 0.00001) populations. Therefore, the pooled OR for the association of GSTM1 null deletions and HCC was calculated using the random-effects model (OR = 1.26, 95% CI: 1.00 - 1.58, P = 0.05). In the subgroup analysis regarding ethnicity, the OR was estimated at 1.32 (95% CI: 1.00 - 1.73, P = 0.05) for Asians and 1.18 (95% CI: 0.78 - 1.78, P = 0.43) for non-Asians. Figure 2 shows the forest plot of these pooled analyses.
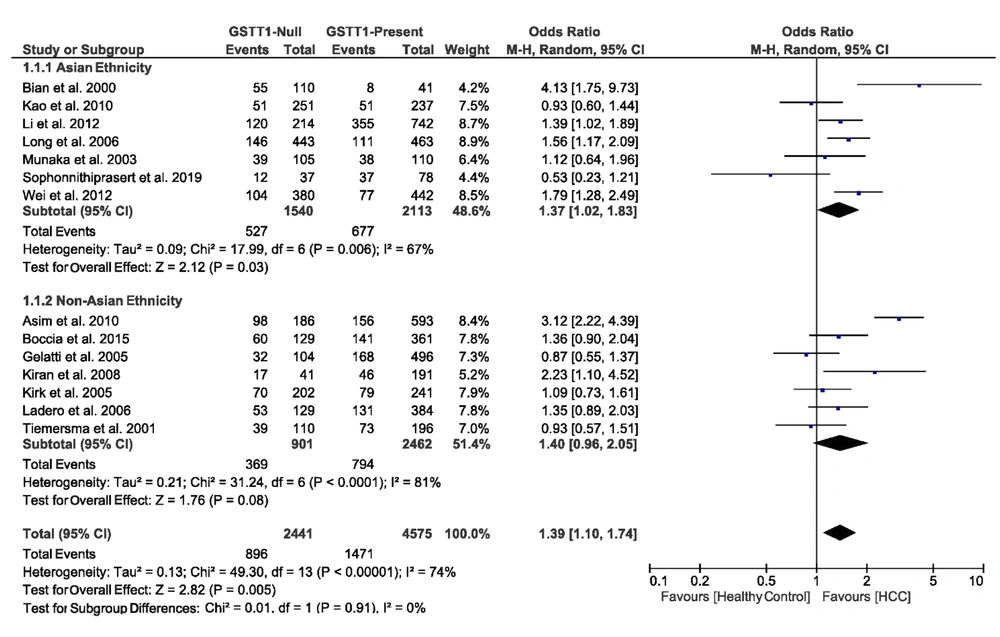
3.4.2. Association of GSTT1 Null Deletions with the Risk of HCC
We found the heterogeneity of data regarding the association of GSTT1 null deletions with the risk of HCC in both Asian (I2 = 67%, P = 0.006) and non-Asian (I2 = 81%, P < 0.0001) populations. Therefore, the pooled OR for the association of GSTT1 null deletions with HCC was calculated in a random-effects model (OR = 1.39, 95% CI: 1.10 - 1.74, P = 0.005). In the subgroup analysis by ethnicity, OR was 1.37 (95% CI: 1.02 - 1.83, P = 0.03) for Asians and 1.40 (95% CI: 0.96 - 2.05, P = 0.08) for non-Asians. Figure 3 demonstrates the forest plot of these pooled analyses.
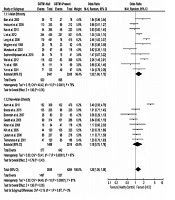
3.4.3. Association of GSTP1 rs1695 Polymorphism with the Risk of HCC
We found heterogeneity of data regarding the association of GSTP1 rs1695 polymorphism with the risk of HCC in both Asian (I2 = 60%, P = 0.08) and non-Asian (I2 = 65%, P = 0.09) populations. Accordingly, the pooled OR for the association of GSTP1 rs1695 polymorphism with HCC was calculated in a random-effects model (OR = 1.14, 95% CI: 0.86 - 1.50, P = 0.36). In the subgroup analysis by ethnicity, the OR was 1.07 (95% CI: 0.75 - 1.54, P = 0.69) for Asians and 1.45 (95% CI: 0.62 - 3.40, P = 0.40) for non-Asians. Figure 4 shows the forest plot of these pooled analyses.
3.5. Publication Bias
According to Figure 5, we found significant publication bias in the evaluated studies.
4. Discussion
Chronic viral hepatitis and exposure to aflatoxins are two main risk factors for HCC in developing countries, making it a major cancer type in these regions (40). The metabolism of aflatoxins varies genetically among different populations, which justifies differences in the prevalence of HCC, despite the similarity of viral hepatitis infection and aflatoxin exposure (41). Therefore, GSTM1 and GSTT1 null deletions and GSTP1 rs1695 polymorphism can predispose people in contact with environmental factors to HCC.
The hemostasis of amino acids (including leucine, isoleucine, and valine) and carbohydrate metabolism are necessary for liver function. All metabolism pathways are controlled by enzymes that are produced by specific genes. Some abnormalities in metabolic pathways, such as redox metabolism, fatty acid metabolism, amino acid metabolism, and drug/hormone metabolism, besides genes involved in these pathways, have been shown to affect the risk and prognosis of HCC (42). By identifying the relationship between the expression level of these genes and the risk of HCC, researchers and physicians can find new methods of treatment and prevention for HCC, according to the individual’s underlying genetic profile.
Phase II enzymes, including those encoded by GSTM1, GSTT1, and GSTP1 genes, play an important role in the detoxification of aflatoxins, as well as other carcinogens (43, 44). Previous research has shown that null genotypes may lead to enzyme deficiency and act as a predisposing factor for HCC (37). However, there are some discrepancies between the results of these studies, and none of them have reached a definite conclusion about the association of GST null genotypes with the risk of HCC; this may be related to the low sample size of these studies or differences in the studied populations and study designs. Accordingly, the present meta-analysis aimed to reach a relatively definite conclusion about the association of GSTM1 and GSTT1 null deletions and GSTP1 rs1695 polymorphism with the risk of HCC.
In the present meta-analysis, 23 studies were included, based on the inclusion criteria. All articles were case-control studies, and most of them were conducted among Chinese and Taiwanese populations. The pooled OR in our meta-analysis showed that GSTM1 and GSTT1 null deletions had significant relationships with the risk of HCC. However, we did not find any significant relationship between GSTP1 rs1695 polymorphism and the risk of HCC. In this regard, Wang et al. (45) conducted a similar study to evaluate the association of GSTT1 and GSTM1 null deletions with the risk of HCC. After assessing 123 studies, 23 papers were included in their meta-analysis; they included studies written in English and Chinese languages. Statistical analysis revealed that independent or concurrent presence of GSTM1 and GSTT1 null deletions significantly increased the risk of HCC in the Asian population, which is consistent with the results of the present study.
In another study, Song et al. (46) evaluated the effects of GSTT1 and GSTM1 null deletions on the risk of HCC. After assessing 287 studies, they finally included 34 articles in their meta-analysis. It should be noted that they included both case-control and cohort studies, which might explain the difference in the number of included studies with the present meta-analysis. The authors concluded that these polymorphisms slightly increased the risk of HCC in the Asian and Indian populations. Additionally, Sui et al. (47), by evaluating the effect of the concurrent presence of GSTM1 and GSTT1 null deletions, reported no direct interaction between these polymorphisms and the risk of HCC; however, each GSTM1 and GSTT1 deletion had its independent impact on the development of HCC. They also concluded that the concurrent presence of these genetic variations had a more significant effect on the risk of HCC in the Chinese population as compared to other populations.
Moreover, a recently published study evaluated the effects of GSTM1 and GSTT1 null deletions on the risk of HCC. Li et al. (5) evaluated 41 articles, including studies written in English and Chinese languages. They reported that these genetic variations increased the risk of HCC in Asians, but not African or Caucasian populations. Nevertheless, this study did not have any precise or united inclusion criteria, which might account for the differences in the number of included studies.
The present study had some limitations. Since we were required to use the data reported in the selected articles, we were unable to perform a meta-analysis in subgroups in terms of gender or age.
In conclusion, the results of the present study suggest significant associations between GSTM1 and GSTT1 null deletions and HCC. However, no significant relationship was found between GSTP1 rs1695 polymorphism and the risk of HCC.