1. Background
1.1. Introduction to Hepatitis C
Hepatitis C is a public health problem worldwide, with around 71 million people infected according to the latest World Health Organization (WHO) estimates (1). Hepatitis C virus (HCV) is one of the leading causes of chronic hepatitis, cirrhosis, and hepatocarcinoma in the world (2).
1.2. Diagnosis of Hepatitis C
Given the suspicion of HCV infection, there are 3 main tests: enzyme immunoassay (EIA) for anti-HCV antibodies, detection of HCV RNA by polymerase chain reaction (PCR), and enzyme-linked immunosorbent assay (ELISA) for HCV core antigen (HCVcAg) (3, 4).
Serology for the detection of antibodies is a very specific test (99%) and currently is the most widely used test for infection screening (5). However, it is not valid to establish an early diagnosis since it presents a window period of 8 - 12 weeks after infection (6).
HCV RNA by PCR is very sensitive and capable of detecting very low viral load (to 10 IU/mL) in as little as 2 days after infection (6).
Another valid method for the diagnosis and monitoring of treatment is to determine the HCVcAg by EIA, detecting it in serum a few days after the HCV RNA is detectable (7). The indications for this determination are the same as for PCR (7).
1.3. Clinical Experience with the Identification of Hepatitis C Virus Core Antigen
HCVcAg is a viral protein released into plasma during viral assembly and, therefore, a marker of HCV replication (8). This antigen has a good correlation with the viral load measured through RNA and can be used for the same indications: screening and diagnosis of acute and chronic infection and reinfection of HCV, being especially useful in monitoring treatment (9).
The study of HCV through an antigenic test offers numerous advantages over PCR (8, 9):
HCVcAg is a more stable protein than RNA; therefore, the test has fewer pre-and post-analytical requirements; also, the methodology for its determination is simple, and it can be performed in any microbiology laboratory, with results available the same day, while PCR requires a week due to the need to batch process samples to be cost-effective (8). This earlier diagnosis would allow treatment to begin earlier and the loss of fewer lives (9).
The main advantage of the antigenic test is the lower cost of the technique (9).
The main limitation of core antigen determination is its lower analytical sensitivity (3 fmol/L, equivalent to 500 - 3000 IU/mL) compared to HCV PCR for the detection of very low viral loads (10). The latter can quantify values above 15 IU/mL and detect as low as 10 IU/mL (11).
Regarding treatment monitoring, the lower sensitivity of HCVcAg concerning RNA occurs at the end of treatment and 4 weeks later, being practically the same at 12 and 24 weeks after finishing treatment (12).
1.4. Work Hypothesis
HCVcAg is a useful and inexpensive technique that can substitute for RNA determination in evaluating the effectiveness of direct-acting antiviral (DAA) treatment in patients with hepatitis C.
2. Objectives
The aim of this study was to see if HCVcAg testing could be used for monitoring the effectiveness of treatment with oral antivirals.
3. Methods
3.1. Participants
A cohort study with retrospective analysis was conducted on patients treated for HCV between July 2014 and February 2020 in the Spanish Hospital of Albacete. Patients over 18 years old diagnosed with HCV infection and treated with DAA with or without cirrhosis, both naïve and pretreated, were included. Each treatment regime was indicated by the relevant experts according to current clinical guidelines and included sofosbuvir / simeprevir (SOF/SIM), ombitasvir / paritaprevir / ritonavir / dasabuvir (OMB/PAR/RIT/DAS) ± SOF, SOF/daclatasvir (DAC), SOF/ledipasvir (LED), SOF/DAC/SIM, SIM/DAC, elbasvir / grazoprevir (EBV/GRZ), SOF / velpatasvir (VEL), glecaprevir / pibrentasvir (GLC/PIB), and SOF / VEL / voxilaprevir (VOX). The duration of treatment was 8, 12, 16, and 24 weeks, with or without adding ribavirin according to their doctor’s choice.
3.2. Ethical Considerations
According to the Declaration of Helsinki, informed consent was obtained from all participants before the initiation of the study. The study protocol was approved by the Clinical Research Ethics Committee of the Puerta de Hierro de Majadahonda University Hospital (Madrid, Spain).
3.3. Variables
The variables were collected in an SPSS database, including demographic characteristics (sex, age), viral genotype, etiology of HCV infection, degree of liver fibrosis, and viral load quantified by both RNA and HCVcAg detection in different moments. In this way, we evaluated HCV RNA levels and core antigen levels at 7 days, 15 days, 4 weeks, and 8 weeks after starting treatment, at the end of treatment, and 4 and 12 weeks after treatment.
The effectiveness of treatment was defined by the presence of undetectable HCV RNA 12 weeks after the withdrawal (sustained virologic response at 12 weeks [SVR12]). HCV RNA was quantified by COBAS TaqMan HCV assay version 2.0 (Roche Diagnostics, Switzerland), with a lower quantification limit of 15 UI/mL and a lower detection limit of 10 UI/mL. Patients with missing SVR12 data were cataloged as non-recovered. HCVcAg levels were measured on aliquots of stored plasma using 2-step chemiluminescent microparticle immunoassay ARCHITECT HCV Ag (Ref. 6L47) on the ARCHITECT-i2000R Immunoassay Analyser (Abbott Diagnostics, Illinois, USA), with a lower detection limit of 3 fmol/L.
3.4. Calculation
Statistical analysis was performed using SPSS version 23 (SPSS Inc, Chicago, Ill, USA). Baseline demographic characteristics were analyzed using frequency measures (absolute and percentages) for qualitative variables, as well as mean, median, and range for quantitative variables. For the comparison of the categorical variables, the Chi-square statistical test was applied. In all cases, differences whose p-value associated with the contrast test was less than or equal to 0.05 were considered significant.
For viral loads, non-parametric statistical tests were performed since they were not normally distributed. The correlation of HCVcAg and HCV RNA was evaluated using the Spearman rho test.
4. Results
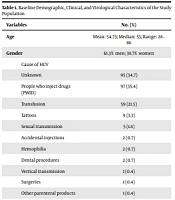
In this study, we included 274 patients. Baseline demographic, clinical, and virological characteristics of the study population are presented in Table 1.
| Variables | No. (%) |
|---|---|
| Age | Mean: 54.73; Median: 53; Range: 28 - 86 |
| Gender | 61.3% men; 38.7% women |
| Cause of HCV | |
| Unknown | 95 (34.7) |
| People who inject drugs (PWID) | 97 (35.4) |
| Transfusion | 59 (21.5) |
| Tattoos | 9 (3.3) |
| Sexual transmission | 5 (1.8) |
| Accidental injections | 2 (0.7) |
| Hemophilia | 2 (0.7) |
| Dental procedures | 2 (0.7) |
| Vertical transmission | 1 (0.4) |
| Surgeries | 1 (0.4) |
| Other parenteral products | 1 (0.4) |
| Genotype | |
| 1b | 117 (43) |
| 1a | 65 (23.9) |
| 3 | 38 (14) |
| 4 | 27 (9.9) |
| Mixed | 14 (5.3) |
| 2 | 4 (1.5) |
| 5 | 1 (0.4) |
| Treatment and SVR12 rates a | |
| OMB/PAR/RIT/DAS | 54 (92) |
| SOF/LED | 48 (93) |
| GLC/PIB | 44 (100) |
| SOF/DAC | 29 (96) |
| SOF/SIM | 21 (95) |
| EBV/GRZ | 18 (94) |
| SOF/VEL | 18 (94) |
| SOF/RVB | 4 (75) |
| SOF/VEL/VOX | 3 (100) |
| Liver fibrosis (kPa) | Mean: 13.185; Median: 8.7; Range: 3.8 - 75 |
| Cirrhotic patients | 90 (32.9) |
| Child | |
| A | 78 (86.63) |
| B | 11 (12.16) |
| C | 1 (1.21) |
a Values are expressed as rate (SVR: %)
At the time of diagnosis, 61.5% of the patients had a high viral load. We considered this when the count of HCV RNA was more than 800 000 IU/mL.
The mean HCV RNA quantification at diagnosis was 2309327 IU/mL, and the median was 1190000 IU/mL with a range of 28 to 22100000 IU/mL. The mean antigenic load was 5972 fmol/L, and the median was 3262 fmol/L; the range was between 0 and 200000 fmol/L. There was a strong correlation between HCVcAg levels and RNA levels with a Spearman rho of 0.832 (P < 0.01).
HCV RNA and HCVcAg load in each time point during treatment and posttreatment follow-up are presented in Table 2.
| Treatment | Viral Load (UI/mL) | Antigenic Load (fmol/L) | ||||
|---|---|---|---|---|---|---|
| Mean | Median | Range | Mean | Median | Range | |
| Pretreatment | 2309327 | 1190000 | 28 - 22100000 | 5972 | 3262 | 0 - 200000 |
| On-treatment (7 days) | 231 | 103 | 10 - 3290 | 2 | 0 | 0 - 16 |
| On-treatment (15 days) | 100 | 19 | 0 - 985 | 1 | 0 | 0 - 17 |
| On-treatment (4 weeks) | 11 | 0 | 0 - 285 | 0 | 0 | 0 - 15 |
| At the end of treatment | 1 | 0 | 0 - 65 | 0 | 0 | 0 - 12 |
| Posttreatment (4 weeks) | 18 | 0 | 0 - 3970 | 0 | 0 | 0 - 5 |
| Posttreatment (12 weeks) | 19084 | 0 | 0 - 9610930 | 103 | 0 | 0 - 16942 |
After the diagnosis of HCV infection and before starting treatment, 262 patients in our study had their HCVcAg determined in turn, obtaining an analytical sensitivity of 99% (95% CI, 97 - 100) when compared with the gold standard.
We evaluated the viral loads of our patients during their treatment and after its completion through HCV RNA and HCVcAg. These results, sensitivity and specificity, and data used for their calculation in each point during treatment and posttreatment follow-up are presented in Table 3.
| Treatment | Samples (n) | True Positive | True Negative | False Positive | False Negative | Sensitivity% (95% CI) | Specificity% (95% CI) |
|---|---|---|---|---|---|---|---|
| Pretreatment | 262 | 259 | 0 | 0 | 3 | 99 (97 - 100) | Not calculable |
| On-treatment (7 days) | 89 | 28 | 1 | 0 | 60 | 32 (21 - 41) | 100 |
| On-treatment (15 days) | 93 | 18 | 22 | 0 | 53 | 25 (15 - 35) | 100 |
| On-treatment (4 weeks) | 261 | 17 | 149 | 11 | 84 | 17 (9 - 24) | 93 (89 - 97) |
| At the end of treatment | 266 | 1 | 241 | 11 | 13 | 7 (( - 8) - 22) | 96 (93 - 98) |
| Posttreatment (4 weeks) | 104 | 1 | 102 | 0 | 1 | 50 (23 - 78) | 100 |
| Posttreatment (12 weeks) | 274 | 5 | 264 | 1 | 0 | 100 | 99 (98 - 100) |
As can be seen in Table 3, at 12 weeks after treatment, we obtained a mean of 19084 IU/mL, a median of 0 IU/mL, and a range of 0 to 9610930 IU/mL for HCV RNA, while for HCVcAg the mean was 103 fmol/L, the median was 0 fmol/L, and the range was 0 to 16 942 fmol/L. At this time point, we also found a strong correlation between HCVcAg levels and HCV RNA levels with a Spearman rho of 0.775 (P < 0.01).
Finally, the virological cure was achieved in 99% of our patients.
After completion of treatment, the sensitivity of the HCVcAg test was 7%, the specificity was 95% (95% CI, 93 - 98%), the positive predictive value (PPV) was 0%, and the negative predictive value (NPV) was 98.72%. However, these values were not statistically significant (P = 0.9).
Four weeks after starting treatment, sensitivity, specificity, PPV, and NPV were 100%.
Regarding the virological cure (SVR12), the sensitivity of the antigenic test was 100%, and the specificity was 99% (95% CI, 98 - 100%). PPV for the HCVcAg test was 88%, while NPV was 100%. The HCVcAg screening test accurately detected all patients with viral recurrence (5/5).
5. Discussion
To achieve the goals set by WHO (1), affordable and effective techniques are urgently needed to diagnose HCV infection, confirm cure, and detect relapses or reinfections. This study supports previous studies, indicating that HCVcAg can be used to identify active virus infection in the initial or screening phase among people with chronic HCV infection, as well as to identify people with SVR12 after completion of treatment (3).
In our study, the analytical sensitivity of HCVcAg at diagnosis was 98%, consistent with previous studies (11).
Further, detection of HCVcAg could be used as a substitute for HCV RNA to diagnose active infection at a lower cost. However, to detect all patients with viremia in a positive anti-HCV sample, if we obtain a negative result for HCVcAg, it would be necessary to confirm it by an RNA assay (12-14). Since a very low number of false negatives with HCVcAg are expected in the general population, the number of RNA tests required to diagnose infection could be reduced by maintaining the detection target of 100% of patients with active-HCV infection. Possibly in Spain, to diagnose all patients infected with HCV, the reference test for viremia screening will continue to be the analytical determination of RNA. However, in other countries with fewer resources, it is an interesting option.
We obtained a strong correlation between viral load and antigen at diagnosis and 12 weeks after treatment. This association is in line with previous studies (15-17).
In our study, the lowest analytical sensitivity occurred at the end of treatment, which there were discrepancies that were not statistically significant; thus, we cannot rule out the possibility that they were due to chance. Sensitivity was 7%, while high specificity and NPV were maintained (98.72% in both). The main limitation of our study is that we obtained very low sensitivity values during treatment monitoring and at the end of it. Thus, we cannot consider HCVcAg determination as an alternative to RNA to assess the follow-up of HCV-infected patients undergoing DAA treatment. It would be necessary to carry out a study with more patients to better assess this point. Another limitation of our study is that the number of samples in different time points is different, which could introduce a selection bias in the comparison of diagnostic performance of HCVcAg across different time points.
Four weeks after treatment, only 1 patient was confirmed positive for HCV viral load using HCV RNA and HCVcAg techniques; our study test reached a sensitivity of 100%. This patient also did not achieve the virological cure. These data suggest that in a patient with persistent viral load 4 weeks after treatment, future treatment failure could be predicted with both laboratory tests. However, numerous studies have demonstrated the limited benefit of viral load assessment at 4 weeks, as well as the low sensitivity of the HCVcAg test regarding HCV RNA at this time point (8, 12).
Regarding the virological cure (SVR12), we only found a discrepancy between HCVcAg and HCV RNA at 12 weeks after treatment in a patient with a confirmed positive antigen (his viral load was confirmed negative by PCR, indicating a false positive). In this same patient, all previous HCVcAg measurements were negative, obtaining only a positive result with a very low viral load (4 fmol/L) when he had achieved the virological cure, measured through HCV RNA. This false positive mainly affected the PPV of the test, obtaining a value of 88.33%, which in other studies it was close to 100% (12).
However, by reassessing the viral load 24 weeks after treatment (the other accepted measure to establish the virological cure), we obtained a negative antigenic result. In the event of a discrepancy between the antigen and RNA, the gold standard must prevail (ie, the determination of the viral load through RNA). On the other hand, we can consider that given a positive result at 12 weeks after treatment, it would be convenient to carry out the analytical test again at 24 weeks before assuming that we are dealing with a patient whose treatment has previously failed because it could be due to a false positive like our case.
Additionally, all patients with a positive result for HCV RNA at 12 weeks were also positive for HCVcAg, thus obtaining a sensitivity of 100%. There were no differences in those patients with low viral load at diagnosis, unlike several other studies in which sensitivity decreased due to this low viral load from the beginning of follow-up (12, 13, 18).
The specificity was always between 93 - 100% (at the end of treatment and 4 and 12 weeks after treatment), consistent with the literature (13, 14, 19, 20).
Therefore, the present study shows that HCVcAg is a valid marker in predicting both therapeutic success and failure defined through SVR12 after DAA treatment.
5.1. Conclusions
The determination of HCVcAg by the EIA technique is as effective as the determination of HCV RNA by PCR in evaluating the response to treatment. This is particularly relevant in lower- and middle-income countries and resource-limited settings where the high cost of labor, equipment, and reagents can prohibit molecular testing.