1. Background
Bile is mainly composed of bile salts, phospholipids, and cholesterol. Bile salt is a kind of sodium salt of the conjugate of bile acid (BA) with either glycine or taurine. The conversion of cholesterol to BAs is closely related to lipid metabolism, fat solute vitamins, and hormones (1). BA levels differ in different liver diseases, and the level of BA in the liver reflects and influences liver function to a certain extent. However, the long-term clinical consequences of high serum BA levels remain unknown.
Hepatic uptake of BA is mainly mediated by sodium-dependent or independent transporters. The sodium taurocholate cotransporting polypeptide (NTCP; gene symbol SLC10A1 in humans) is a major transporter of conjugated BA from the serum compartment into hepatocytes (2, 3). Besides, NTCP also transports additional substrates, such as thyroid hormones and drugs (4, 5).
Mutations in the SLC10A1 gene, whose cytogenetic location is on chromosome 14 (14q24.1), may alter the expression and/or function of the NTCP protein, thus influencing bile salts transportation (6-10). The single nucleotide variant (p.Ser267Phe/c.800C>T) of NTCP/SLC10A1 is associated with high levels of BA. The minor allele frequency (MAF) of SLC10A1S267F in East Asia was reported to be between 8% and 12% (9, 11, 12). The SLC10A1S267F variant exhibits a near-complete loss of function for bile salt uptake; yet, it possesses a fully normal transport function for the non-BA substrates such as estrone sulfate (8). This indicates that Ser267 may be placed in a protein region specific for BA transport. Therefore, any clinically impactful changes in the SLC10A1S267F variant may affect liver detoxification or increase susceptibility to hepatotoxicity in patients.
According to recent references in which only a total of 48 individuals, including 38 children and 10 adults (13-19), have been reported as SLC10A1S267F homozygous patients, the mutation of SLC10A1S267F is probably related to elevated serum BAs (a condition often referred to as hypercholanemia). However, the biochemical description of this mutation and whether it is the only reason influencing BAs in patients remain unclear. In this study, we identified 5 SLC10A1S267F homozygous patients and investigated the clinical manifestations and long-term consequences. Here, we also analyze if there exists a combination of multiple gene mutations to influence serum BA levels in SLC10A1S267F patients.
2. Methods
2.1. Human Subject
A total of 12 Han Chinese individuals were recruited at the First Affiliated Hospital of Army Medical University. We identified that 5 individuals with high serum levels of BAs were SLC10A1S267F homozygous, while the other 7 individuals were heterozygous (n = 3) and wild-type (n = 4). We collected basic clinical information and peripheral blood samples from the individuals. Lab examinations, including liver and renal function tests, serum lipids level tests, and routine blood tests, were performed for each subject. This study was approved by the Ethics Committee of the First Affiliated Hospital of Army Medical University, PLA (People's Liberation Army of China) (code: KY2021116). All the study subjects or their legal guardians signed written informed consent.
2.2. Whole-Genome Sequencing
We performed a whole-genome sequencing (WGS) analysis of 12 individuals in this study. Using TruSeq Nano DNA LT Sample Preparation Kits (Illumina Inc, San Diego, CA, USA), the libraries were constructed. Briefly, using an S220 Focused-Ultrasonicator (Covaris, USA), the genomic DNA was sheared into fragments with a length of ~350 base pairs (bp). Using an Illumina sequencing platform, a HiSeq X Ten platform (Illumina Inc, San Diego, CA, USA), the final libraries were sequenced after polymerase chain reaction (PCR) amplification and purification, and 150-bp paired-end reads were generated. The raw sequence data were processed using the NGS-QC toolkit (20) for quality control of sequence data. The clean reads were aligned to the reference genome (GRCh38/hg19) using the Burrows-Wheeler Aligner (21) (BWA), and the average coverage is 29.34X. After alignment, Picard was employed to mark duplicate reads, and SAMtools (22) was used to transform the reads into compressed binary alignment map (BAM) files for sorting and indexing. The genome analysis toolkit (GATK) (23) were used to verify the accuracy of single nucleotide polymorphisms (SNP) and insertion and deletion (InDel). Finally, ANNOVAR (24) was used to perform annotation for the variants.
2.3. Gene Variation Screening
After completing the annotation and filtering steps, we focused on genes known to code for cholesterol and BA metabolism. Firstly, we used “bile acid” and “cholesterol” as keywords and selected all genes coding for human cholesterol metabolism, especially for BA metabolism, using the UniProt database and obtained a total of 560 genes. We further retrieved the SNP file from the WGS results and obtained 709 SNPs (except for SLC10A1S267F) from the recruited subjects. Because homozygotes have greater phenotype and clinical importance, we decided to narrow down the number of SNPs by excluding the heterozygous mutation, meaning that those SNP genotypes should be homozygous in individuals 1 to 5. A total of 560 genes were filtered and reduced to 62 genes covering 76 missense variants. Secondly, to evaluate benign or deleterious of those variants, we searched for their clinical significance in ClinVar or from previous studies and classified them as benign or deleterious variants. Meanwhile, for mutations not previously reported, we used MAF < 0.05 in the public database of 1000 genomes project for the East Asian population and protein prediction tools, including sorting tolerant from intolerant (SIFT; dbNSFP version 3.3a) and polymorphism phenotyping version 2 (Polyphen 2 version 2.2), to forecast the underlying functional impacts. The protein function prediction of SIFT is based on the SIFT score calculated by sequence homology and the physical/chemical similarity between amino acids (deleterious means sift ≤ 0.05; tolerated means sift > 0.05). On the other hand, we used the polyphen 2 HDIV score for protein function prediction, which is based on the HumanDiv database for complex diseases (probably damaging: score ≥ 0.957; possibly damaging: Score = 0.453 ~ 0.956; benign: score ≤ 0.452). Besides, we examined the pathogenesis of the variants reported in all related studies. There were 31 possibly damaging variations that might be related to hypercholanemia (Table 1).
| Gene | rs | Variation | Clinical Significance | MAF | Functional Effects a | Genotype of Individual 1/2/3/4/5/6/7/8/9/10/11/12 | |
|---|---|---|---|---|---|---|---|
| ABCA12 | rs7560008 | c.T1375A | p.S459T | Benign | 0.0016 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| ABCB11 | rs2287622 | c.T1331C | p.V444A | Progressive familial intrahepatic cholestasis | > 0.01 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HE/HO |
| ABCC2 | rs927344 | c.A116T | p.Y39F | Dubin - Johnson syndrome | 0.0054 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| ADGRG6 | rs1262686 | c.A3296G | p.Q1099R | Benign | 0.0004 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| AKR1C4 | rs4880718 | c.A749G | p.Q250R | / | 0.0002 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| APOB | rs679899 | c.C1853T | p.A618V | Familial hypercholesterolemia, Familial hypobetalipoproteinemia | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| APOB | rs584542 | c.A6937G | p.I2313V | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HE/HO |
| APOL1 | rs2239785 | c.G394A | p.E132K | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HE/HO/HE |
| APOL1 | rs136175 | c.G630A | p.M210I | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HE/HO/HE |
| ATP11C | rs2491014 | c.T342G | p.C114W | / | 0.0079 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| ATP8B1 | rs222581 | c.G3454A | p.A1152T | / | 0.0002 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| CD44 | rs1467558 | c.T689C | p.I230T | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| CTAGE5 | rs7140561 | c.T17C | p.V6A | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HE/HE/WT/HE/HE/HE |
| CTAGE5 | rs1950952 | c.G991C | p.E331Q | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| CUBN | rs2796835 | c.C8150G | p.S2717W | Benign | 0.0000 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| CUBN | rs1276712 | c.G6485A | p.C2162Y | Benign | 0.0066 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| DBT | rs12021720 | c.A1150G | p.S384G | Benign | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| EGF | rs4698803 | c.A2633T | p.E878V | Benign | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| FCGBP | rs782538403 | c.G3650A | p.R1217Q | / | 0.0020 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| FCGBP | rs782416774 | c.A3700G | p.T1234A | / | 0.0048 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| FCGBP | rs782342257 | c.G3707A | p.R1236Q | / | None | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| GC | rs9016 | c.A1334G | p.H445R | / | 0.0072 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| LIPC | rs6083 | c.A644G | p.N215S | Hepatic lipase deficiency | > 0.01 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| LIPC | rs3829462 | c.C1068A | p.F356L | Hepatic lipase deficiency | > 0.01 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HE/HE/HO/HE/HO |
| PCSK9 | rs562556 | c.G1420A | p.V474I | Familial hypercholesterolemia, Familial hypobetalipoproteinemia | > 0.01 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| PEX16 | rs10742772 | c.G346A | p.V116I | Benign | 0.0000 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| PROM2 | rs12992066 | c.A1523G | p.Q508R | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HE/HO/HO/HO/HE/HE |
| SLC14A1 | rs11877062 | c.C10T | p.R4W | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HE/HE/HO |
| TF | rs2692696 | c.A1342G | p.I448V | / | 0.0068 | Tolerated | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| TTC39B | rs1407977 | c.A1051G | p.I351V | / | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
| VPS33B | rs11073964 | c.G1459A | p.G487S | Arthrogryposis with renal dysfunction and cholestasis syndrome | > 0.01 | Deleterious | HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO/HO |
Variations in Bile Acids and Cholesterol Metabolism Genes, Detected by Whole - Genome Sequencing
2.4. Sanger Sequencing Validation
Sanger sequencing was used to confirm the SLC10A1S267F mutation in all recruited individuals. PCR primers were designed and used to amplify the 309/419-bp fragment covering the SLC10A1 gene (forward primer, 5’-GCTAGAAACTTGCTTGTTG-3'; reverse primer, 5’-CTCTGAGTGTATGTGGGGT-3’ or forward primer, 5’-GAGTGCAGTTGATGGAAGT-3’; reverse primer, 5’-CTGAGTGTATGTGGGGTTT-3’). The SLC10A1 gene information was obtained from the National Center for Biotechnology Information database.
2.5. Statistical Analysis
Data were analyzed using SPSS version 23 (SPSS Inc, Chicago, Ill, USA). Continuous variables with normal distribution were presented as mean ± SEM; non-normal variables were reported as median (interquartile range). A comparison of the clinical laboratory results among the 3 study groups (wild-type, heterozygote, and homozygote) was analyzed by either parametric or non-parametric tests (1-way analysis of variance [ANOVA] or Kruskal-Wallis omnibus test) as appropriate. P values less than 0.05 were considered statistically significant.
3. Results
3.1. Description of the Phenotype
We recruited a total of 12 subjects and divided them into 3 groups; homozygote, heterozygote, and wild-type (Figure 1; Table 2). Telephone follow-ups revealed good cognition competence and a normal social ability for both adults and children. Adults of homozygotes and heterozygotes had well-developed secondary sexual characteristics and were fertile. Consistent with previous studies (13-19), the level of serum BA was much higher in homozygotes than in wild-type individuals (P = 0.027), meaning that all 5 homozygotes had hypercholanemia, whereas heterozygotes and wild-type individuals had normal serum BA levels. Many factors could affect serum BA levels, needing to be fully excluded before diagnosing SLC10A1S267F mutation-induced hypercholanemia. Thus, a comprehensive medical history was obtained from each homozygote to determine any other diseases that could lead to high levels of serum BA. However, we did not find any individuals with symptoms of cholestatic liver diseases/malabsorptive diseases or long-term medication history.
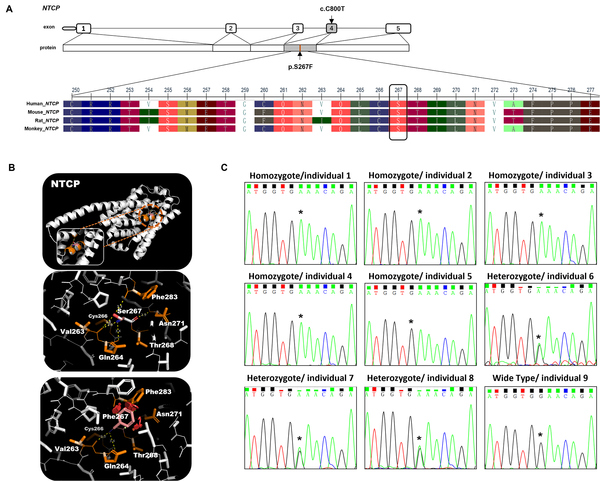
Bioinformatic analysis of the SLC10A1 gene mutation. A, schematic view of NTCP exons and multiple sequence alignment information of NTCP; B, three - dimensional structures, prediction of NTCP wild - type and mutation; C, the Sanger sequencing results of the SLC10A1 gene in 9 individuals. Individuals 1 to 5 were homozygous for the SLC10A1 variants c.800c> T (p.Ser267Phe), individuals 6 to 8 were heterozygous for the SLC10A1 variants c.800c> T (p.Ser267Phe), and individual 9 was wild - type for the SLC10A1 variants c.800c> T (p.Ser267Phe). Individuals 10 to 12 were not included in this analysis due to the limited peripheral blood sample.
| Variables | Homozygotes | Heterozygotes | Wild-Type | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| Sex | Male | Male | Female | Female | Female | Female | Male | Female | Female | Male | Female | Male |
| Age (y) | 36 | 2 | 11 | 7 | 5 | 36 | 33 | 31 | 9 months | 43 | 44 | 17 |
| TBA (umol/L) | 33.1 | 7 | 34 | 35.2 | 47 | 1.9 | 4.9 | 287.7 | 2.8 | 3.6 | 1.7 | 9.1 |
Clinical Characteristics of Individuals
We compared liver and renal functions between the 3 groups (Table 3). The levels of biochemical markers of liver injury were normal, except for high alkaline phosphatase activity in children because of the accelerated growth. The globulin levels decreased but not significantly. Also, the A/G ratio increased in the homozygotes (P = 0.036) and heterozygotes (P = 0.039) but with no clinical phenotype. The level of total bilirubin was lower in homozygotes than in wild-type individuals (P = 0.014) and heterozygotes but still within the normal range; the decreased total bilirubin might influence iron absorption in the long term in homozygotes.
| Genotype (rs2296651) | Wild-Type (GG) (n = 4) | Heterozygous (GA) (n = 3) | Homozygous (AA) (n = 5) | ||
|---|---|---|---|---|---|
| Test | Ref. | Unit | |||
| Liver/Renal Function | |||||
| ALT | 0 - 42 | (IU/L) | 14.87 ± 2.76 | 21.00 ± 18.25 | 20.00 ± 11.77 |
| AST | 0 - 42 | (IU/L) | 17.70 (17.65, 23.35) | 16.00 (15.50, 21.50) | 30.00 (26.00, 42.00) |
| ALP | 34 - 114 | (IU/L) | 192.33 ± 199.95 | 59.33 ± 32.32 | 190.20 ± 90.73 |
| GGT | 5 - 50 | (IU/L) | 13.67 ± 3.21 | 20.67 ± 9.07 | 20.67 ± 7.20 |
| TBA | 0 - 10 | (umol/L) | 2.80 (2.25, 3.20) | 4.90 (3.40, 5.95) | 35.20 (34.00, 47.00) b |
| TBIL | 6 - 21 | (umol/L) | 12.70 ± 3.93 | 11.03 ± 1.37 | 6.82 ± 2.17 b |
| DBIL | 0 - 6 | (umol/L) | 2.57 ± 0.67 | 4.50 ± 1.84 | 2.98 ± 0.89 |
| TP | 66 - 83 | (g/L) | 72.83 ± 0.90 | 72.30 ± 1.93 | 70.40 ± 5.69 |
| Alb | 38 - 51 | (g/L) | 46.60 (45.40, 47.20) | 45.20 (45.0, 46.6) | 49.00 (48.80, 49.00) |
| G | 25 - 38 | (g/L) | 26.63 ± 2.72 | 24.00 ± 5.19 | 20.84 ± 4.09 |
| A/G | 1.2 - 2.5 | 1.76 ± 0.25 | 1.77 ± 0.17 b | 2.45 ± 0.49 b | |
| UA | 155 - 428 | (umol/L) | 374.67 ± 124.64 | 286.33 ± 138.60 | 277.80 ± 87.42 |
| Urea | 1.7 - 8.3 | (mmol/L) | 3.97 ± 1.07 | 3.81 ± 0.63 | 3.12 ± 1.26 |
| Cre | 45 - 84 | (umol/L) | 61.40 (46.20, 61.6) | 43.00 (42.50, 57.35) | 36.00 (21.00, 38.00) |
| Blood Fat | |||||
| TCH | 3.1 - 5.7 | (mmol/L) | 4.63 ± 0.30 | 4.00 ± 1.14 | 4.16 ± 0.90 |
| TG | 0.4 - 1.73 | (mmol/L) | 1.28 (1.02, 2.185) | 0.76 (0.53, 1.56) | 1.07 (0.78, 1.43) |
| HDL - C | 0.9 - 2 | (mmol/L) | 1.49 ± 0.52 | 1.44 ± 0.39 | 1.52 ± 0.31 |
| LDL - C | 2.07 - 3.1 | (mmol/L) | 2.73 ± 0.28 | 2.36 ± 0.81 | 2.64 ± 0.91 |
| Serum Thyroid Hormone | |||||
| T3 | 1.3 - 3.1 | (nmol/L) | 1.35 (1.30, 1.58) | 1.70 (1.65, 1.76) | 2.09 (1.86, 2.16) |
| T4 | 66 - 181 | (nmol/L) | 98.70 (94.41, 121.22) | 91.43 (89.87, 94.35) | 110.05 (104.74, 144.10) |
| FT3 | 3.1 - 6.8 | (pmol/L) | 2.97 ± 0.70 | 2.57 ± 0.09 | 3.28 ± 0.52 |
| FT4 | 12 - 22 | (pmol/L) | 15.82 (15.69, 16.55) | 16.49 (14.96, 17.09) | 21.49 (18.22, 23.09) |
| TSH | 0.27 - 4.20 | (uIU/mL) | 0.78 ± 0.33 | 1.01 ± 0.39 | 0.56 ± 0.19 |
| Vitamin D | |||||
| Vitamin D (25(OH)D) | 25 - 200 | (pmol/L) | 50.60 (47.18, 55.02) | 45.06 (42.86, 47.55) | 32.26 (30.60, 34.57) |
| Routine Blood Tests | |||||
| WBC | 3.5 - 9.5 | (109/L) | 7.78 ± 1.00 | 5.69 ± 0.64 | 7.48 ± 2.56 |
| NEU | 1.8 - 6.3 | (109/L) | 4.47 ± 0.81 | 3.33 ± 0.71 | 2.62 ± 0.88 b |
| LYM | 1.1 - 3.2 | (109/L) | 2.79 ± 0.30 | 1.79 ± 0.10 b | 3.97 ± 2.74 |
| MON | 0.1 - 0.6 | (109/L) | 0.35 ± 0.16 | 0.39 ± 0.01 | 0.51 ± 0.21 |
| BAS | 0 - 0.06 | (109/L) | 0.00 (0.00.0.11) | 0.01 (0.005, 0.01) | 0.02 (0.01, 0.03) |
| EOS | 0.02 - 0.52 | (109/L) | 0.13 (0.09.0.21) | 0.20 (0.12, 0.23) | 0.15 (0.05, 0.17) |
| RBC | 4.3 - 5.8 | (1012/L) | 4.88 ± 0.33 | 4.95 ± 0.50 | 4.79 ± 0.56 |
| HGB | 115 - 150 | (g/L) | 141.00 ± 9.64 | 141.00 ± 24.02 | 127.80 ± 17.58 |
| MCV | 82 - 100 | (%) | 86.57 ± 5.26 | 88.13 ± 12.63 | 86.04 ± 13.52 |
| MCH | 27 - 34 | (pg) | 28.93 ± 1.87 | 28.43 ± 4.87 | 26.92 ± 3.71 |
| MCHC | 316 - 354 | (g/L) | 333.00 (333.00, 335.00) | 318.00 (313.00, 331.00) | 311.00 (309.00, 317.00) |
| RDW - SD | 37 - 54 | (%) | 41.20 ± 1.01 | 46.07 ± 3.56 b | 43.58 ± 1.82 |
| RDW - CV | 11 - 16 | (%) | 13.00 (12.75, 13.50) | 13.70 (13.25, 14.95) | 14.10 (14.00, 14.40) |
| PLT | 125 - 350 | (109/L) | 287.00 ± 51.47 | 195.67 ± 17.67 b | 310.40 ± 25.15 c |
| MPV | 9 - 13 | (fl) | 9.70 ± 0.26 | 9.47 ± 1.02 | 9.66 ± 0.79 |
| PDW | 9 - 17 | (fl) | 11.50 ± 0.10 | 16.13 ± 0.90 b | 15.66 ± 0.52 b |
Biochemical Parameters of the Study Cohort Grouped by Genotype a
Apart from the liver and renal functions, we also determined the levels of blood lipids (including total cholesterol, triglyceride, high-density lipoprotein cholesterol [HDL-C], and low-density lipoprotein cholesterol [LDL-C]), thyroid hormones (T3, T4, FT3, FT4, TSH), and vitamin D (25(OH)D). Previous studies (13, 14, 16) have reported that patients with a homozygous SLC10A1S267F mutation were prone to vitamin D deficiency and abnormal blood lipid levels. However, in our study, all homozygotes had normal blood lipids, thyroid hormones, and vitamin D levels (Table 3). Compared to heterozygotes and wild-type subjects, in homozygotes, some indices (such as T3, T4, FT3, and FT4) increased while TSH decreased; however, all parameters were within the reference range.
The clinical chemistry parameters are presented in Table 1. The neutrophil count in homozygotes decreased (P = 0.015), whereas the lymphocyte and monocyte counts were slightly elevated. Some biochemical markers, including platelet (PLT) and platelet distribution width (PDW) in homozygotes, significantly differed compared with the heterozygotes (P = 0.001) and wild-type subjects (P < 0.001), respectively–but with a small clinical difference (Table 3). Interestingly, in heterozygotes, the lymphocyte count and PLT significantly decreased (P = 0.046 and P = 0.09), and PDW increased (P < 0.01). Taken together, these findings suggest that individuals with or without an SLC10A1S267F mutation are healthy. However, the small differences in results should be studied further.
3.2. Validation of the <i>SLC10A1</i><sup>S267F</sup> Mutation Using WGS and Sanger Sequencing
WGS and Sanger sequencing were performed to confirm the presence of the SLC10A1S267F mutation in the subjects. Individuals 1 to 5 were homozygotes, individuals 2 to 7 were heterozygotes at the variation allele, and individuals 9 to 12 were wild-type (Figure 1B). The Sanger sequencing results were consistent with WGS results.
3.3. Relationship of the <i>SLC10A1</i><sup>S267F</sup> Mutation and Other Gene Mutations in Homozygotes
We noted that in some cases, SLC10A1S267F homozygotes with a combination of other gene mutations or diseases showed different symptoms, such as jaundice and/or liver dysfunction, late fetal loss, or urticarial rashes (13, 15, 17, 25, 26). We confirmed that serum BA levels increased asymptotically in SLC10A1S267F homozygotes. Approximately 90% of cholesterol would be converted into BA by a series of enzymes and reactions in the liver. Therefore, cholesterol metabolism-related gene mutations could affect BA metabolism and might influence serum BA levels. To confirm if other variants could influence serum BA levels in homozygotes, we performed further gene analysis on these individuals. A total of 62 genes (coding for cholesterol metabolism and/or BA metabolism) covering 76 SNP were selected. The ClinVar public database and MAF were used to evaluate those variants. We also used protein prediction tools to predict underlying functional impacts in metabolism and find out deleterious variants. After filtering, 31 possibly damaging SNP were identified (see Table 1). According to references and prediction tools, except for the SLC10A1S267F mutation, there are 5 SNPs that have been thought to be the most possible factors in influencing serum BA levels in recruited homozygotes (ABCB11V444A, ABCC2Y39F, APOBA618V, PSCK9V474I, and VPS33BG487S). None of the previous studies about SLC10A1S267F have mentioned these SNPs, and SLC10A1S267F in homozygotes was thought to be the most likely cause of high BA levels. Although we did not figure out the relationship between the 5 SNPs and hypercholesterolemia in this study, these variations will be studied in our future research.
4. Discussion
Non-synonymous variants of SLC10A1, including c.668T>C(p.Ile223Thr), c.800C>T(p.Ser267Phe), c.836T>C(p.Ile279Thr), c.940A>G(p.Lys314Glu), and c.755G>A(p.Arg252His), have been reported previously. The p.Ser267Phe variant exhibits a loss of function for bile salt uptake, but it keeps a fully normal transport function for the non-BA substrate.
In this study, a biochemical assessment of recruited individuals was done to enrich the characterization of the SLC10A1S267F mutation, which is more comprehensive compared with previous studies. Bioinformatics and clinical evidence in this study suggested that this variant was none-pathogenic, which is consistent with previously reported cases of NTCP deficiency. However, the neutrophil count in 2 children (individuals 3 and 8) was lower than normal, while their lymphocyte count was above normal. Two children with a homozygous SLC10A1S267F mutation also presented decreased globulin, suggesting that researchers and doctors should focus on the immunologic function of homozygotes and their susceptibility to pathogens.
Bile and bile salts are vital for endogenous compounds and metabolic products, such as the digestion of dietary fat and fat-soluble vitamins (27-30). According to previous studies, fat-soluble compounds (such as vitamin D) deviate from the normal range in SLC10A1S267F homozygotes. However, our patients displayed slight changes in vitamin D and serum lipid levels. Thus, the specific molecular role of SLC10A1S267F in fat solubilization and absorption needs to be further elucidated.
Some studies have mentioned that patients with SLC10A1S267F display worse clinical phenotype (such as liver dysfunction), always accompanied by other damage variants. Also, our previous results indicated that Slc10a1S267F homozygous mice displayed normal levels of serum BA and did not cause hypercholanemia (26). Thus, to our knowledge, there are probably other variants in SLC10A1S267F homozygotes; in this regard, 5 candidate SNPs are described below.
ABCB11 and ABCC2 are genes coding for the bile salt export pump (BSEP) and multidrug resistance-associated protein 2 (MRP2), respectively. BSEP and MRP2 are bile salt transporters located at the canalicular membrane of hepatocytes (6). BSEP mainly participates in hepatic BA and lipid homeostasis. In most cases, mutations in BSEP are closely related to inherited progressive cholestatic liver disorders in infancy, known as progressive familial intrahepatic cholestasis (PFIC) (31, 32). Missense variant ABCB11V444A is related not only to PFIC but also to a risk factor of intrahepatic cholestasis in pregnancy or drug-induced liver injury (33-35). Therefore, we speculate that 444Ala constitutes a susceptibility factor for the development of hypercholanemia under certain conditions, such as a combination mutation of SLC10A1S267F. MRP2 can transport divalent bile salts. The ABCC2Y39F mutation has found in SLC10A1S267F homozygotes. Some mutations of single amino acid in ABCC2 could confer MRP2 new transport capacity for bile salts, indicating that we should pay attention to this SNP and its underlying effect on serum BA levels.
Mutations of APOBA618V and PSCK9V474I were reported as related factors of familial hypercholesterolemia in the ClinVar database. Apolipoprotein B (APOB) is a major protein constituent of chylomicrons (apo B-48), LDL (apo B-100), and VLDL (apo B-100) (36). The APOBA618V mutation is associated with higher LDL-C levels (odds ratio [OR] = 1.41) (37), meaning that cholesterol and BA levels in SLC10A1S267F homozygotes could be influenced to some extent. The coexistence of APOBA618V and APOBA618V should be studied. The main function of PCSK9 is the degradation of the LDL receptor. In some cases, PCSK9 increases apoB100 production, reduces APOB degradation to increase circulating LDL levels and contributes to hypercholesterolemia and atherosclerosis (38). However, PSCK9V474I seems to be a protection variant associated with lower LDL and cholesterol (39). This might be the reason why the serum lipids in SLC10A1S267F homozygotes were normal in this study.
VPS33B is a class C vacuolar protein sorting protein (40). Approximately 75% of arthrogryposis, renal dysfunction, and cholestasis syndrome (ARC) patients harbor germline mutations in VPS33B. ARC is characterized by congenital joint contractures, renal tubular acidosis, and neonatal jaundice, leading to death in infancy. However, in milder cases, patients survive into adulthood and increase serum BA values but normal bilirubin levels (41). The VPS33BG487S mutation in SLC10A1S267F homozygotes is predicted as a non-specific risk factor in ARC. However, the relationship between VPS33BG487S and SLC10A1S267F mutation still needs to be studied.
From the above discussion, we suggest that although the SLC10A1S267F mutation is the most important reason for high serum BA levels, we detected some mutations in homozygotes that might influence BA metabolism. Further studies on these variants could provide a deeper understanding of the etiology of hypercholesterolemia.
Although we provided a comprehensive clinical, biochemical explanation of the SLC10A1S267F mutation and discussed how to explore the etiology of the SLC10A1S267F mutation-related hypercholesterolemia, there are some limitations. Due to experimental conditions and equipment limitations, we only used medical history data to exclude cholestasis/ malabsorptive diseases or long-term medication history (such as anti-hyperlipidemic agents). Also, we did not evaluate the insulin resistance status of the subjects.
Apart from the function of transporting molecules, NTCP has other functions. Convincing human genetic evidence supports the role of NTCP as a cellular receptor for hepatic B virus (HBV). Recently, several epidemiology studies have reported SLC10A1S267F homozygotes with lower HBV entry and hepatocyte infection compared to wild-type individuals. SLC10A1S267F homozygotes may escape from HBV infection and present decreased cholesterol levels as a consequence of impaired BA uptake (42). According to the marked higher allele frequency of p.S267F in Asia, this SNP might be a positive selection of Asian for its advantage in conferring resistance to chronic HBV. We aim to design more in vivo and in vitro experiments to study and analyze these variants of SLC10A1 in the next phase of our investigation. We have already constructed Slc10a1S267F mutation mice and prepared for further study to determine whether there are deleterious effects on the population and how 267Phe influences HBV infection. To understand the etiology of hypercholanemia, a mouse model of a Slc10a1S267F mutation combined with other related variants is also needed. The effect of p.Ser267Phe on other BA transport proteins should also be explored.
4.1. Conclusions
This study presents an in-depth characterization of NTCP p.Ser267Phe mutations in 5 homozygotes by clinical examination, genetic analysis, and functional prediction. The results supported that the p.Ser267Phe mutation has no deleterious clinical effects in both children and adults.