1. Background
By the advent of targeted therapies and its initial success in inducing a profound impact on cancer patients outcome, a new gate of hope has been opened for patients to overcome the disease; nonetheless, significant challenges are still remained to maximize the clinical benefit of targeted therapy (1). Ever-increasing attempts have been done thus far to identify more novel biomarkers not only to predict the prognosis but also to target it through therapeutic interventions (2). Among different types of human cancers, leukemia, in particular acute myeloid leukemia (AML), is notorious for its heterogeneous genetic features as multiple molecules and signaling pathways play fundamental roles in the molecular pathogenesis of this malignancy (3). Moreover, despite the promising improvements in the treatment strategies and significant increase in 5-year event-free survivals (EFS), still the early incidence of relapse in AML, which is widely associated with the aberrant expression of mounting body of genes, is a frequent event (4). Notably, this unique characteristic put AML at the forefront of the priority to be fully studied for the identification of novel biomarkers that are involved in the primary stages of leukemogenesis (5).
In the pathogenesis of AML, deregulation of several signaling pathways has been reported to participate in the maintenance of hematopoietic progenitor cells survival. Since PI3K signaling axis is located at the upstream of survival pathways and oncogenic mediators, several studies have reported that the inhibition of this axis, either as a single treatment or in combination with chemotherapy, could be a valuable strategy to eliminate the survival of leukemic cells (6-10). However, although this inhibition was quite effective, the precise role of the PI3K network in the pathogenesis of AML is still a debatable issue. Although there are some reports suggesting that the activation of PI3K could participate in the early stage of AML formation, others claimed that the aberrancy in this pathway is acquired after the application of chemotherapeutic drugs (11). Apart from direct impact on the evolution of myeloid leukemia cells, the regulatory role of the PI3K pathway on the expression of c-Myc as an oncogene with a well-established influence on the transformation of myeloid progenitors has resulted in re-consideration of PI3K role in the pathogenesis of AML (12). The discrepancies on the pathogenic value of PI3K and its downstream targets in AML patients on one hand, and the importance of the proposing a plausible mechanism underlying AML pathogenesis on the other hand, encouraged us not only to evaluate the expression levels of the main component of the PI3K pathway including Akt, c-Myc, CIP2A, and PP2A in newly diagnosed AML patients but also to investigate if there is any correlation between aforementioned genes in this leukemia.
2. Objectives
Given the involvement of the variety of signaling pathways foremost the PI3K axis in the pathogenesis of human cancers, we aimed at investigating the expression of the most important downstream targets of this pathway to propose a plausible mechanism underlying AML pathogenesis.
3. Methods
3.1. Patients Sample Collection
From October 2018 to August 2019, peripheral blood samples obtained from 30 de-novo AML patients and 10 healthy controls. The research protocol was approved by the Research Ethics Committee at the Shahid Beheshti University of Medical Sciences (ID number: IR.SBMU.RETECH.REC.1397.1012) and all participants signed informed consent document in accordance with the statement of Helsinki. The clinical characteristics of participants with de novo AML are illustrated in Table 1. Patients were diagnosed according to French-American-British (FAB) classification, which categorizes AML subtypes based on morphologic characteristics of malignant cells. Among all collected samples, 86.5% were diagnosed as non-M3 AML and 13.5% were categorised as AML-M3. The mean age of the patients at the time of diagnosis was ~56 years. Noteworthy, 56% of the samples (16 out of 30) were collected from male and 44% of the samples (14 out of 30) were allocated to female patients.
| No. | FAB | RBC × 106 | WBC × 103 | Hb (g/dL) | Hct (%) | Plt × 103 | Blast (%) |
|---|---|---|---|---|---|---|---|
| 1 | AML-M2 | 3.32 | 24.7 | 9.7 | 28.4 | 48 | 35 |
| 2 | AML-M1 | 2.79 | 50.01 | 9.2 | 27.9 | 165 | 50 |
| 3 | AML-M2 | 3.12 | 86.45 | 9.5 | 28.1 | 78 | 65 |
| 4 | AML-M2 | 4.41 | 32.14 | 12.9 | 38.7 | 170 | 40 |
| 5 | AML-M0 | 2.98 | 45.24 | 10.1 | 28.8 | 93 | 35 |
| 6 | AML-M4 | 2.71 | 23.39 | 8.7 | 26.1 | 238 | 25 |
| 7 | AML-M2 | 3.58 | 14.46 | 10.5 | 29.3 | 24 | 60 |
| 8 | AML-M5 | 3.5 | 37.06 | 9.8 | 31.6 | 55 | 25 |
| 9 | AML-M3 | 2.95 | 75.54 | 9.2 | 25.7 | 53 | 45 |
| 10 | AML-M4 | 3.53 | 11.11 | 10.6 | 31.3 | 80 | 80 |
| 11 | AML-M2 | 5.34 | 15.34 | 14.6 | 42.1 | 160 | 45 |
| 12 | AML-M3 | 5.19 | 56.61 | 15.8 | 46.0 | 137 | 55 |
| 13 | AML-M1 | 2.31 | 108 | 6.01 | 21.0 | 10 | 40 |
| 14 | AML-M2 | 4.1 | 36.45 | 11.39 | 34.0 | 78 | 80 |
| 15 | AML-M4 | 5.01 | 13 | 14.6 | 42.14 | 175 | 80 |
| 16 | AML-M5 | 3.03 | 36.54 | 9.5 | 27.3 | 45 | 90 |
| 17 | AML-M3 | 2.42 | 99.14 | 8.3 | 24.8 | 141 | 43 |
| 18 | AML-M4 | 4.18 | 16.36 | 12.3 | 38.05 | 189 | 26 |
| 19 | AML-M6 | 4.45 | 17.45 | 13.5 | 40.4 | 173 | 32 |
| 20 | AML-M2 | 3.56 | 54.04 | 11.6 | 34.4 | 121 | 41 |
| 21 | AML-M1 | 3.06 | 14.4 | 9.3 | 26.6 | 63 | 21 |
| 22 | AML-M2 | 2.75 | 29.5 | 10.3 | 31.4 | 69 | 38 |
| 23 | AML-M4 | 3.43 | 27.76 | 9.5 | 31.3 | 91 | 41 |
| 24 | AML-M2 | 3.37 | 106.7 | 9.4 | 29.2 | 35 | 68 |
| 25 | AML-M3 | 3.11 | 19.45 | 9.3 | 27.7 | 116 | 51 |
| 26 | AML-M2 | 3.33 | 16.86 | 10.3 | 30.2 | 31 | 65 |
| 27 | AML-M4 | 4.17 | 18.54 | 9.8 | 28.1 | 26 | 35 |
| 28 | AML-M5 | 3.67 | 21.41 | 8.6 | 26.1 | 32 | 40 |
| 29 | AML-M2 | 4.02 | 13.52 | 11.7 | 34 | 15 | 30 |
| 30 | AML-M1 | 2.89 | 42.51 | 9.9 | 29.41 | 62 | 28 |
Abbreviations: Hb, hemoglobin; RBC, red blood cells; WBC, white blood cells.
3.2. RNA Purification and Preparation of cDNA
Total cellular RNA from mononuclear cells (MNCs) of participants was extracted using High Pure RNA isolation kit (Qiagen, Germany) as recommended by the supplier. The amount of extracted RNA was spectrophotometrically assessed using Nanodrop ND-1000 (Optical Density (OD) 260/280 nm ratio > 1.8). Subsequently, 1 µg RNA was used to synthesize cDNA in a final volume of 20 µL using a Thermo fermentas kit according to the manufacturer’s protocol (Thermo scientific - USA).
3.3. Real-Time Quantitative PCR
To evaluate the expression levels of genes, sense and antisense primers of target genes were designed by GeneRunner software (details are shown in Table 2). Alterations in the mRNA expression levels of genes were measured using SYBR green-based real-time quantitative polymerase chain reaction (qRT-PCR) (Rotor-Gene 6000, Qiagen, Germany). A 15 µL reaction containing 2 μL cDNA, 1 μL Sense and antisense primers, 7.5 μL of master Mix (Ampliqon, Denmark), as well as 4.5 μL water was amplified in thermal cycler. For each target gene, primer efficiency was estimated from a standard curve using four consecutive 1:10 dilutions of the cDNA sample. ABL was amplified as housekeeping genes. All tests were done in triplicate and fold changes in the expressions of each mRNA were calculated using the Livak method (2-∆∆CT) (13).
| Gene | Accession Number | Forward Primer (5’-3’) | Reverse Primer (5’-3’) | Size |
|---|---|---|---|---|
| ABL | NM-005157 | TCCTCGTCCTCCAGCTGTTA | ACCCGGAGCTTTTCACCTTT | 218 |
| Akt | NM-005163 | TCCTCCTCAAGAATGATGGCA | GTGCGTTCGATGACAGTGGT | 181 |
| c-Myc | NM-002467 | CCACAGCAAACCTCCTCACAG | GCAGGATAGTCCTTCCGAGTG | 105 |
| CIP2A | NM-020890 | ACCCCAACATAAGTGCTTCAC | TCTCTGGTTTCAATGTCTACTGC | 79 |
| PP2A | NM-178001 | TCTCAGGCATACGCTGACTAC | GGAGACTCTGTACTCGAAGGT | 90 |
3.4. Statistical Analysis
All the statistical analyses were performed using the SPSS software (version 16.0) and the GraphPad Prism 6 software. Independent student t or Mann-Whitney U tests were conducted for comparison between patients and the control group. Pearson’s correlation test was used to study the possible correlation between the indicated genes. A probability value of less than 0.05 was considered statistically significant.
4. Results
4.1. The Expression Level of CIP2A and PP2A in AML Samples
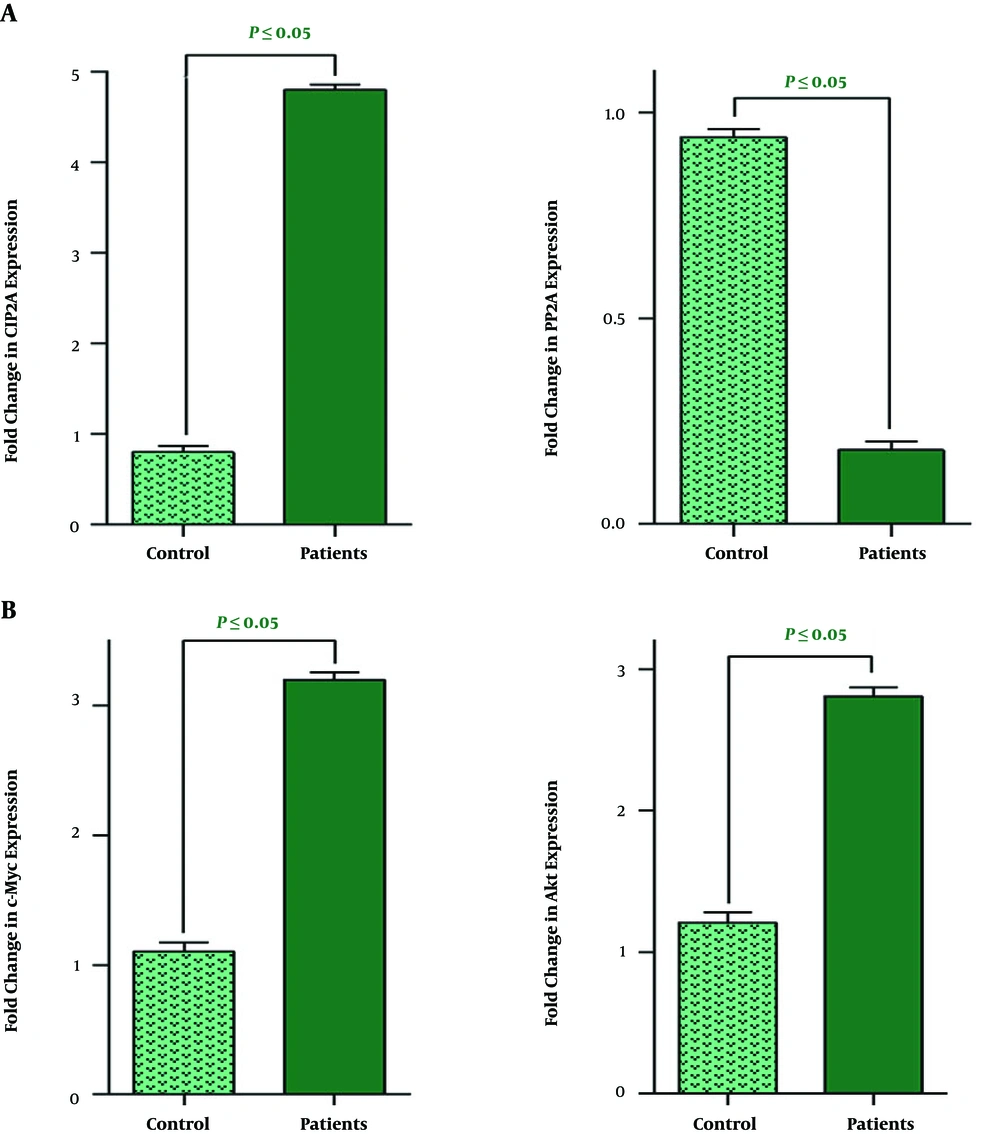
The prognostic value for CIP2A, a direct inhibitor of a tumour suppressor PP2A, has been reported in wide range of human solid tumours (14); however, its association with human leukaemia has not yet been clarified. To assess the expression level of CIP2A and its downstream target PP2A in AML and also to explore its plausible contributory role in leukomogensis, peripheral blood of 30 AML patients (56% male and 44% female) with the median age of 49 years old were collected for quantitative real-time PCR analysis. In addition, 10 normal cases were also included as a control group in order to provide a better outlook for the alteration in the gene expression pattern. We found that the increased expression of CIP2A in AML samples was coupled with the reduction in mRNA level of its downstream target PP2A (Figure 1A). Based on the results, the expression level of both CIP2A and PP2A was different in AML patients and healthy individuals, it was of particular interest to evaluate whether the alteration in the expression pattern of these genes has any association with age, gender, and the percentage of blasts. As presented in Table 3, we found the similar expression pattern both in male and female groups and also in all age categories that we have divided our samples into. The same result has been also obtained when we compared the mRNA level of these genes with the percentage of the blasts (Table 3).
| Clinical Variables | Total Patients, 30 (100) | Evaluable Patients | Evaluable Patients | ||||
|---|---|---|---|---|---|---|---|
| CIP2A (↑) | CIP2A (↓) | P Value | PP2A (↑) | PP2A (↓) | P Value | ||
| Age, year | 49, 19 - 83 | 0.325 | 0.871 | ||||
| n < 60 | 19 (63.3) | 14 (46.6) | 5 (17) | 7 (23.3) | 12 (40) | ||
| n ≥ 60 | 11 (36.4) | 8 (26.4) | 3 (10) | 3 (10) | 8 (26.7) | ||
| Sex | 0.470 | 0.312 | |||||
| Male | 17 (56) | 10 (33.3) | 7 (23.4) | 4 (13.3) | 13 (43.3) | ||
| Female | 13 (44) | 9 (30) | 4 (13.3) | 3 (10) | 10 (33.4) | ||
| Percentage of blast | 0.556 | 0.117 | |||||
| n < 40 | 10 (33.3) | 9 (30) | 1 (3) | 2 (6) | 8 (26.7) | ||
| n ≥ 40 | 20 (66.7) | 13 (43.3) | 7 (23.7) | 4 (13.3) | 16 (54) | ||
| AML subtype | 0.281 | 0.120 | |||||
| M3 | 4 (13.5) | 2 (6.5) | 2 (6.5) | 1 (3) | 3 (10) | ||
| Non-M3 | 26 (86.5) | 20 (67) | 6 (20) | 5 (16.4) | 21 (70.6) | ||
Abbreviation: AML, acute myeloid leukemia.
aValues are expressed as No. (%) or median, range.
bStatistically significant values: P < 0.05.
4.2. The Expression Level of Akt and c-Myc Was Increased in AML Patients as Compared to Healthy Counterparts
Previous studies have shown that there is a cross-talk between PP2A downregulation and c-Myc expression in malignant cells (15). It has been reported that not only c-Myc could suppress the expression of PP2A and thereby prolong the survival signals in the neoplastic cells, but also the decrease in the expression level of PP2A is associated with stabilizing phosphorylated c-Myc in malignant cells (16). Given this and based on our results, we also aimed to examine the expression level of c-Myc in AML patients. The results of the qRT-PCR analysis revealed that the expression level of this oncogene was significantly higher as compared with the healthy counterparts (Figure 1B); suggestive of the oncogenic role of c-Myc in the leukemogenesis process. It has been reported that c-Myc, located at the downstream of the PI3K/Akt signaling pathway, serves as a mediator to transmit the survival signals to the nucleus and thereby orchestrate the variety of cellular functions (17). Accordingly, the results of qRT-PCR analysis indicated that the expression level of the Akt has been elevated in the AML samples as compared to its expression in the healthy individuals (Figure 1B). By applying the Livak method, we also found that there was not a significant correlation between the mRNA expression patterns of these genes with age, gender, and the percentage of blasts (Table 4).
| Clinical Variables | Total Patients, 30 (100) | Evaluable Patients | Evaluable Patients | ||||
|---|---|---|---|---|---|---|---|
| Akt (↑) | Akt (↓) | P Value | c-Myc (↑) | c-Myc (↓) | P Value | ||
| Age, year | 49, 19 - 83 | 0.254 | |||||
| n < 60 | 19 (63.3) | 13 (43) | 6 (20) | 0.435 | 15 (50) | 4 (13.3) | |
| n ≥ 60 | 11 (36.4) | 9 (30) | 2 (15) | 7 (23.4) | 4 (13.3) | ||
| Sex | 0.120 | ||||||
| Male | 17 (56) | 12 (40) | 5 (16.6) | 0.740 | 11 (36.6) | 6 (20) | |
| Female | 13 (44) | 10 (33.3) | 3 (10) | 10 (33.3) | 3 (10) | ||
| Percentage of blast | 0.391 | ||||||
| n < 40 | 10 (33.3) | 7 (23.3) | 3 (10) | 0.276 | 8 (26.3) | 2 (6) | |
| n ≥ 40 | 20 (66.7) | 15 (50) | 5 (16.7) | 16 (53.3) | 4 (13.4) | ||
| AML subtype | 0.726 | ||||||
| M3 | 4 (13.5) | 3 (10) | 1 (3) | 0.178 | 3 (10) | 1 (3) | |
| Non-M3 | 26 (86.5) | 22 (73.3) | 4 (13.7) | 20 (67) | 6 (20) | ||
Abbreviation: AML, acute myeloid leukemia.
aValues are expressed as No. (%) or median, range.
bStatistically significant values: P < 0.05.
4.3. Correlation Between Akt, c-Myc, CIP2A, and PP2A Expression Levels in the Studied Groups
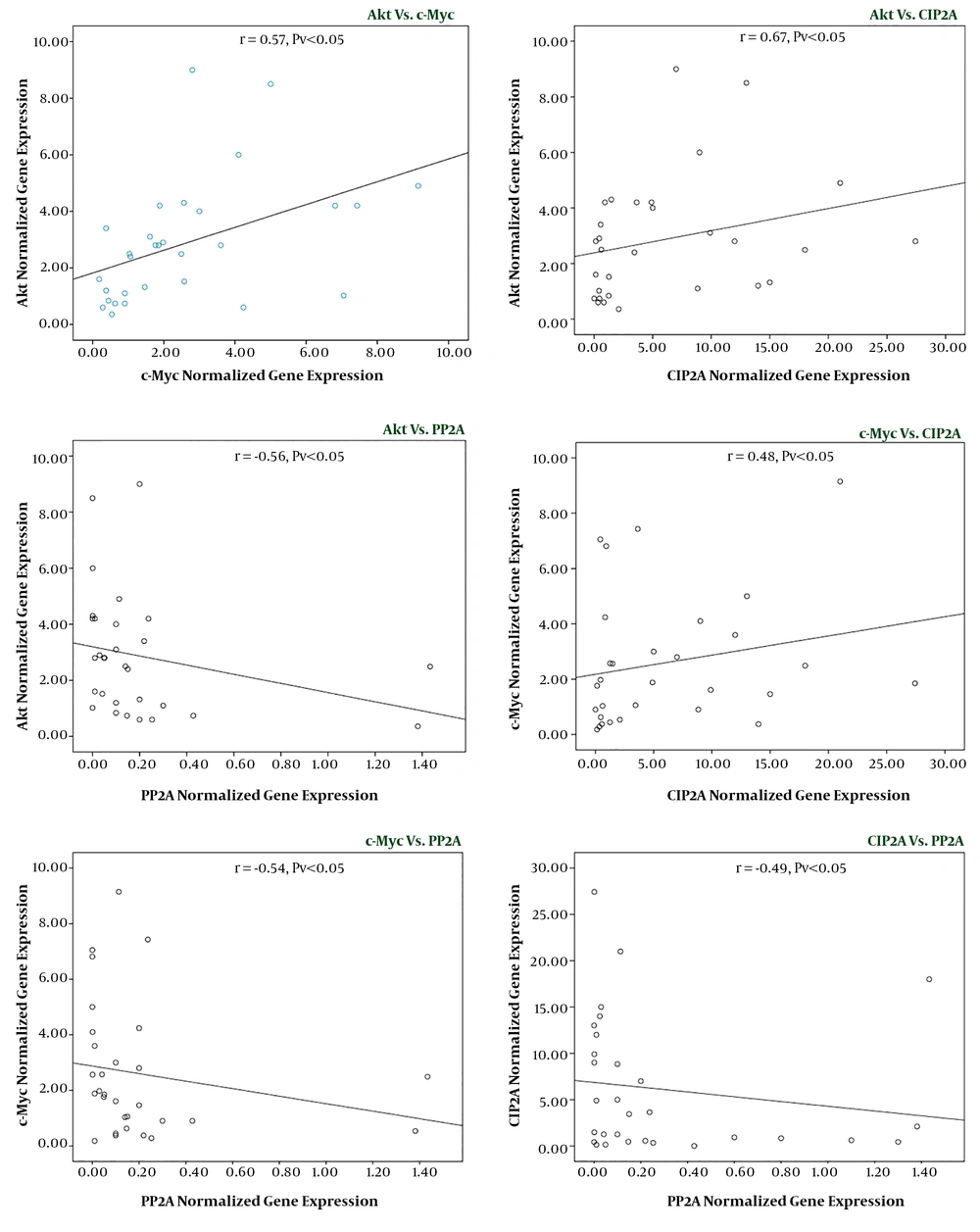
Having established that there was a significant difference between the expression levels of the indicated genes among AML patients and healthy counterparts, we then used statistical correlation analysis to evaluate whether there is a significant correlation between the expression levels of the genes in the patients group. Our results showed that a significant correlation exists between gene expression levels of PP2A and CIP2A (r = -0.49, P < 0.01), Akt and c-Myc (r = 0.57, P < 0.05), c-Myc and PP2A (r = -0.54, P < 0.01), Akt and PP2A (r = -0.65, P < 0.01), c-Myc and CIP2A (r = 0.48, P < 0.05), and Akt and CIP2A (r = 0.67, P < 0.05) (Figure 2); showing the prominent role of this axis in the pathogenesis of AML.
A, while the expression levels of CIP2A in leukemic patients were significantly higher than the control group, the mRNA expression level of PPI2A significantly decreased; B, there was a significant elevation in the expression levels of both Akt and c-Myc in leukemia patients as compared with samples collected from the normal volunteers. Values are given as mean ± standard deviation of three independent experiments.
Correlation between the expression level of Akt, c-Myc, CIP2A, and PP2A in leukemic samples. Normal distribution of data was achieved by log 10 computation and correlation between Akt, c-Myc, CIP2A, and PP2A was determined in 30 AML patients. Values are given as mean ± standard deviation of three independent experiments.
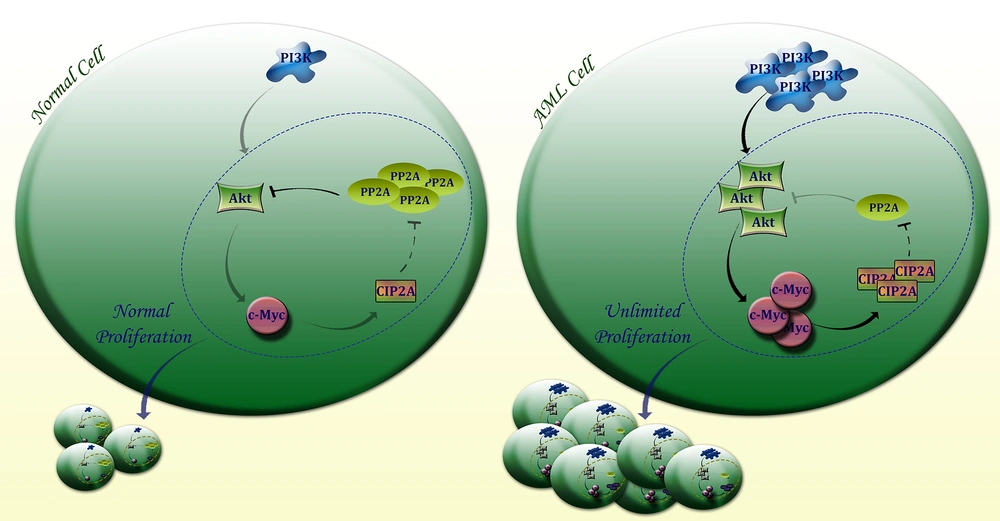
Schematic representation proposed for the plausible role of PI3K axis in AML cells. Through aberrant activation of the PI3K signaling pathway which subsequently leads to the over-expression of Akt, c-Myc would be expressed in malignant cells and turn lead to the excessive expression of CIP2A, a direct inhibitor of PP2A. Upon PP2A suppression, Akt and c-Myc remained phosphorylated and this defective loop may be responsible for providing an opportunity for AML cells to proliferate more vigorously.
5. Discussion
Despite dramatic therapeutic advances in recent years, the minimal overall survival (OS) for AML patients ascertained the importance of the identification of the new biomarkers to provide a better outlook of the disease pathogenesis (18). When the results of genetic and molecular experiments showed that the aberrancy in the expression or activity of c-Myc oncogene is a frequent occurrence in AML patients, intense interest has been attracted to this factor to evaluate its association with either pathogenesis or prognosis of the disease. The results of the mutation analysis in AML patients showed that in contrast to several malignant hematopoietic cells such as multiple myeloma (MM) and non-Hodgkin lymphoma (NHL), which in them c-Myc serves both as prognostic and pathogenic mediators (19, 20), mutations in c-Myc gene may not play a pivotal role in the initiation of AML (21). However, Xia et al. revealed that c-Myc may serve as a resistance biomarker in AML patients (22). In the present study, we found that the expression level of c-Myc was elevated in AML samples; however, we could find no significant correlation between its expression and the difference in age, gender, and the percentage of the blasts. In agreement, accumulating evidence also indicated that the high expression level of c-Myc in the neoplastic myeloid cells could be an important criterion for the development of the disease and the induction of drug-resistance in AML patients (23, 24). Notably, the results of other investigations showed that by inhibition of c-Myc in acute leukemia cells, not only the survival of the cells were significantly reduced (25, 26), but also this suppression resulted in the enhancement of the therapeutic value of chemotherapeutic drugs (26, 27), which demonstrates the contributory role of c-Myc in the pathogenesis of AML.
Multiple mediators have been demonstrated to be involved in the regulation of c-Myc in malignant cells, which amongst them the PI3K signaling pathway has been recognized as one of the most important axis to harness the activity of this oncogene (28). Apart from c-Myc regulation, given the harmonizing role of the PI3K axis in the wide range of intracellular functions, the oncogenic role of this network has always been under the magnifying glass. Analysis of the crucial role of the PI3K aberrancy and more precisely the over-expression of Akt, as the main downstream component of the PI3K axis, in chronic lymphoid leukemia (CLL) recently led to the application of the PI3K inhibitors in the therapeutic approaches of this malignancy (29), which in turn resulted in an increased tendency to study the role of this axis in the other types of hematologic malignancies. Some studies suggested that in different types of AML, AML-M3 (APL) which is notorious for the translocation of PML-RARα displays a greater amount of over-activated Akt (30, 31). Accordingly, other studies indicated that the cell lines of AML-M3 are more sensitive to the antileukemic effect of PI3K inhibitors as compared to other AML-derived cell lines (32). However, in the present study, we found that in AML patients either diagnosed as M3 or non-M3, the expression level of Akt was greater than healthy counterparts, which was in agreement with the elevated c-Myc in these patients; further highlighting the fact that the activation of the PI3K/Akt/c-Myc axis may be a plausible event participating in AML pathogenesis.
Genomic and proteomic approaches have widely declared that the sync between PI3K and c-Myc would not be accomplished unless PP2A phosphatase would be inhibited by its dominant inhibitor CIP2A (cell proliferation regulating inhibitor of PP2A) (33). In another word, whether through PI3K or c-Myc, CIP2A would be activated and subsequently by the suppression of PP2A this oncogene would construct a defective loop through which both Akt and c-Myc would be stabilized, giving the malignant cells an opportunity to proliferate more vigorously (33, 34). In agreement, our results delineated that in AML patients with an over-expressed profile of both c-Myc and Akt, there was an elevation in the expression level of CIP2A. Interestingly, we also found that the mRNA level of PP2A was decreased in AML patients as compared to the control group. The role of PP2A in AML pathogenesis has not yet been well-clarified, and this study for the first time suggested that probably this gene serves as a missing point through which PI3K/c-Myc axis could regulate the expression level of CIP2A (Figure 3).
5.1. Conclusions
Taken together and as the most straightforward interpretation of our results, we proposed that probably the association between PI3K and c-Myc which is built through the interaction between CIP2A and PP2A may play a pivotal role in the pathogenies of AML and any component of this axis could serve as a potential new target to interfere in the novel treatment strategies. However, further detailed investigations in this field are required to clarify the exact role of this interesting testis-specific pathway in the context of hematological malignancies, in particular AML.