1. Background
Osteosarcoma (OS), known as osteogenic sarcoma, is an aggressive and primary bone tumor with an incidence rate of 4 to 5 cases per million, affecting children and adolescents (1-5). More importantly, it accounts for 19% of all malignant tumors of bone (6) with a higher prevalence in young males compared with young females (5), and its etiology is still opaque (4, 7). It affects the metaphysis of long bones, such as the distal femur, and it seems that rapid bone growth occurring in adolescents is related to the promotion of malignancy (8). Although there is still a controversy about the origin of OS, accumulative evidence has shown that it is regarded as a differentiation disease caused by genetic changes, leading to the disruption of the differentiation of mesenchymal stem cells into osteoblast (1). OS, as heterogenic cancer, is characterized by broad ranges of the genomic instability, increased abnormality in karyotypes, and multiple genomic aberrations, including copy number gains and losses in several chromosomes (2). Calin et al. demonstrated, for the first time, the relationship between the susceptibility of various cancers and miRNAs in 2002 (9, 10).
Recent studies have indicated that non-coding RNAs, such as microRNA with conserved sequences exert essential roles in both normal physiological processes, including the development, differentiation, metabolism, and proliferation, as well as disease progression that include malignant tumors (2, 7, 10, 11). Several lines of evidence show that the dysregulation of miRNAs is associated with the initiation and disease progression of tumors, along with their invasion, metastasis, and chemo-resistance; however, their contribution to the development of OS is still unclear (7, 10, 12). MiRNAs mainly consist of 22 nucleotides involved in RNA silencing by post-transcriptional regulation of gene expression, changing the stability of specific mRNAs, or obstructing the translation of some mRNAs. Hence, any alteration in the expression of miRNAs may influence the expression of tumor-suppressor and oncogene genes (4, 9, 13-16). Gao et al. studied the role of miRNA expression in the development of OS (2). MiR-135b plays a functional role in the normal differentiation of osteoblasts (1, 17, 18). Also, the presence of miR-135b in the placenta is capable of regulating the growth rate of its cells (19). It has been revealed that miR-135b contributes to the differentiation of mesenchymal cells into bone and muscle cells during embryonic development (20). Moreover, the decreased expression of miRNAs is necessary for the normal mineralization of osteoblast cells (16).
Recent data suggest that FOXO-1, one of the FOXO family members of the transcription factors, participates in a variety of biological events, such as cell proliferation, apoptosis, stress response, and DNA damage response. Several studies have shown that FOXO-1 is involved in many types of cancers (21-23). There is strong evidence indicating that the upregulated expression of c-Myc is a major factor in the initiation of OS (23, 24). c-Myc, as an oncogene gene, is overexpressed in many types of cancers, and it contributes to cell migration, cell invasion, metastasis, and cell cycle (25). On the other hand, although FOXO expression is frequently reduced in numerous types of cancers, the increased expression of the transcription factor c-Myc has been further reported (26). c-Myc, as a transcription factor, is a pivotal player in different cellular processes, including cell migration, growth, and proliferation (27, 28).
In the case of cancer, the expression of miR-135b is impaired, resulting in cancer cell promotion, migration, and invasion (20). Mechanistically, its oncogenic function is mediated via the repression of some crucial components of the Hippo pathway, which controls the organ size in Drosophila melanogaster and tumorigenesis in mammals (19). Although a large proportion of the available data agrees with the oncogenic role of miR-135b, its function in the context of cancer is highly controversial. For instance, it has been shown that miR-135b promotes several types of cancers, such as breast and lung cancers (29, 30). However, contradictory results have demonstrated that miR-135b suppresses the invasion of prostate cancer cells and abrogates the chemoresistance of lung cancer cells (31, 32).
2. Objectives
The current research aimed at evaluating the precise role of miRNA-135b in the development of OS, thereby determining the targets of miR-135b and their impact on cell proliferation, cell invasion, and cell migration.
3. Methods
3.1. Sample Collection
In this study, 21 OS tissue samples, along with 21 adjacent normal bone tissues, as control specimens were obtained from patients with a definite diagnosis of OS. Patients were referred to the Shefa Hospital (Tehran, Iran) between 2015 and 2018. The collected tissue samples were stored in liquid nitrogen at -80°C until the analysis.
3.2. Cell Culture
The Saos-2 cell-line was procured from the cell bank of the Culture Collection of Pasteur Institute (Tehran, Iran). The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Carlsbad, CA, USA) that was supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) and penicillin/streptomycin antibiotics (Gibco, Carlsbad, CA, USA). The cells were incubated in an incubator with 5% CO2 at 37°C.
3.3. Cell Transfection
MiR-135b inhibitor, miR-135b mimic, and the controls were provided by the BIONEER company (Daejeon, South Korea). The concentration of miRNAs employed in this research was 20 nmol/L. The process of the cell transfection was facilitated by polyethyleneimine (PEI) that compacts the DNA molecule into particles with positive charges, leading to the binding of the desired molecules to anionic cell surface residues and uptake of them by the cell membrane under a cellular phenomenon called endocytosis. The experiments were carried out in triplicate.
3.4. Real-Time PCR
The miRNA contents of the cultured cells and isolated tissues were extracted, using the miRNeasy Mini kit (QIAGEN, Cat.No.217004). The extracted miRNAs were applied for the synthesis of complementary DNA (cDNA), using the miScript II RT Kit (QIAGEN, Cat.No.218161). The synthesized cDNA samples were used for the amplification of the target genes, using the miScript SYBR Green PCR Kit (QIAGEN, Cat.No.218061). The primers used for the analysis of the expression of miR-135b, as well as the endogenous control RNU6, were obtained from Tag Copenhagen A/S (Denmark). A miRNA-specific forward primer and a universal reverse primer were applied for the subsequent qPCR step. The sequences of the forward primers used in our study were as follows; miR-135b: 5’-CGTATGGCTTTTCATTCCTATGT-3’, U6: 5’-CGCAAGGATGACACGCAA ATT C-3’, and the sequence of the miScript Universal Primer in the miScript SYBR Green kit is proprietary. The relative expression of miRNAs was determined according to the ∆∆CT method.
3.5. Cell Proliferation Assay
For the comparison of the rate of cell proliferation rate between untransfected cells and transfected cells, the MTT assay was utilized. In brief, 15 × 103 cells per well were seeded onto a 96-well microplate and incubated for 24 hours. Next, the cell transfection was conducted, using PEI; then the cells were incubated for 48 hours. Afterward, the cell culture medium was exchanged with the 10% MTT solution (5 mg/mL thiazolyl blue tetrazolium bromide in PBS; Sigma. Aldrich), and the cells were incubated at 37°C for 4 hours. Consequently, the reaction was terminated by the addition of 100 µL DMSO to dissolve the produced formazan crystals. The optical absorbance of the samples was read at a wavelength of 570 nm, using a microplate reader (Biotek Instruments, Inc. Winooski, VT, USA).
3.6. Cell Invasion Assay
For the comparison of the invasiveness between untransfected cells and transfected cells, the Cell Invasion Assay Kit (Biovision) was utilized according to the manufacturer’s recommendations. In brief, 24 hours after the transfection process, cells were seeded on the upper chamber, where the ECMatrix TM gel was pre-coated for 24 hours. Afterward, 700 µL of the 10% FBS-containing medium was supplemented with the lower chamber as a chemoattractant agent. The cells were, then, incubated at 37°C in 5% CO2 for 48 hours. Next, non-invading cells were removed, using a swab, while cells migrated into the bottom of the membrane were fixed with 3.7% formaldehyde. Next, the cells were permeabilized by 100% methanol and, finally, stained with the Giemsa solution. For the quantification of the assay, stained cells were counted in multiple fields under a light microscope, and the mean numbers of the cells were reported.
3.7. Cell Migration Assay
The cell migration technique was conducted according to the scratch test. In brief, 24 hours after the transfection procedure, mitomycin C was added to the culture media to prevent further cell division. After 2 hours, an artificial gap was produced, using a 100 µL pipette tip (scratching). Then, the cell culture medium was exchanged with fresh DMEM that was devoid of fetal bovine serum. Regularly, cells on the edge of the created gap tend to move toward the groove to fill-in the gap until new cell-cell contact is established. For the visualization of the cell migration process, images were taken at 0 and 24 hours. The ImageJ software (NIH, Bethesda, USA) was employed for the calculation of cell migration.
3.8. Western Blot
For the extraction of the protein contents of the cells, transfected cells were lysed through radioimmunoprecipitation assay buffer (RIPA). Afterward, the concentration of the protein content of each sample was determined, using the BCA Protein Assay Kit (Thermo Fisher Scientific Inc. USA). The equivalent amount of extracted proteins were loaded on 10% SDS-PAGE and electrophoretically separated and, then, transferred onto a polyvinylidene difluoride (PDVF) membrane (Amersham, Buckinghamshire, UK). The PVDF membrane was blocked by 5% skim milk in Tris-buffered saline containing 0.1 % Tween 20 for 1 hour. Next, the membrane was incubated with specific primary antibodies against c-MYC and FOXO-1 proteins (Rabbit monoclonal antibodies, Cell Signaling Technology, Danvers, MA, USA) overnight. The membrane was, then, washed 3 times and incubated with HRP-conjugated goat anti-rabbit IgG secondary antibody (Cell Signaling Technology, Danvers, MA, USA) at room temperature for 1 hour. The probed antibodies were, then, visualized by the ECL detection reagent (Roche, Basel, Switzerland) and, subsequently, the signal intensity of the protein bands was quantified, using the ImageJ software (NIH, Bethesda, USA).
3.9. Statistical Analysis
The statistical analysis was conducted by the SPSS software version 21. Data were expressed as the means ± standard deviation (mean ± SD). The differences between the two experimental groups were analyzed by the Student’s t test, while for multiple comparisons, one-way analysis of variance (ANOVA) was utilized, followed by Tukey’s post hoc test. The P value of less than 0.05 was statistically considered significant and each experiment was performed at least 3 times.
4. Results
4.1. Increased Level of miR-135b Expression in OS
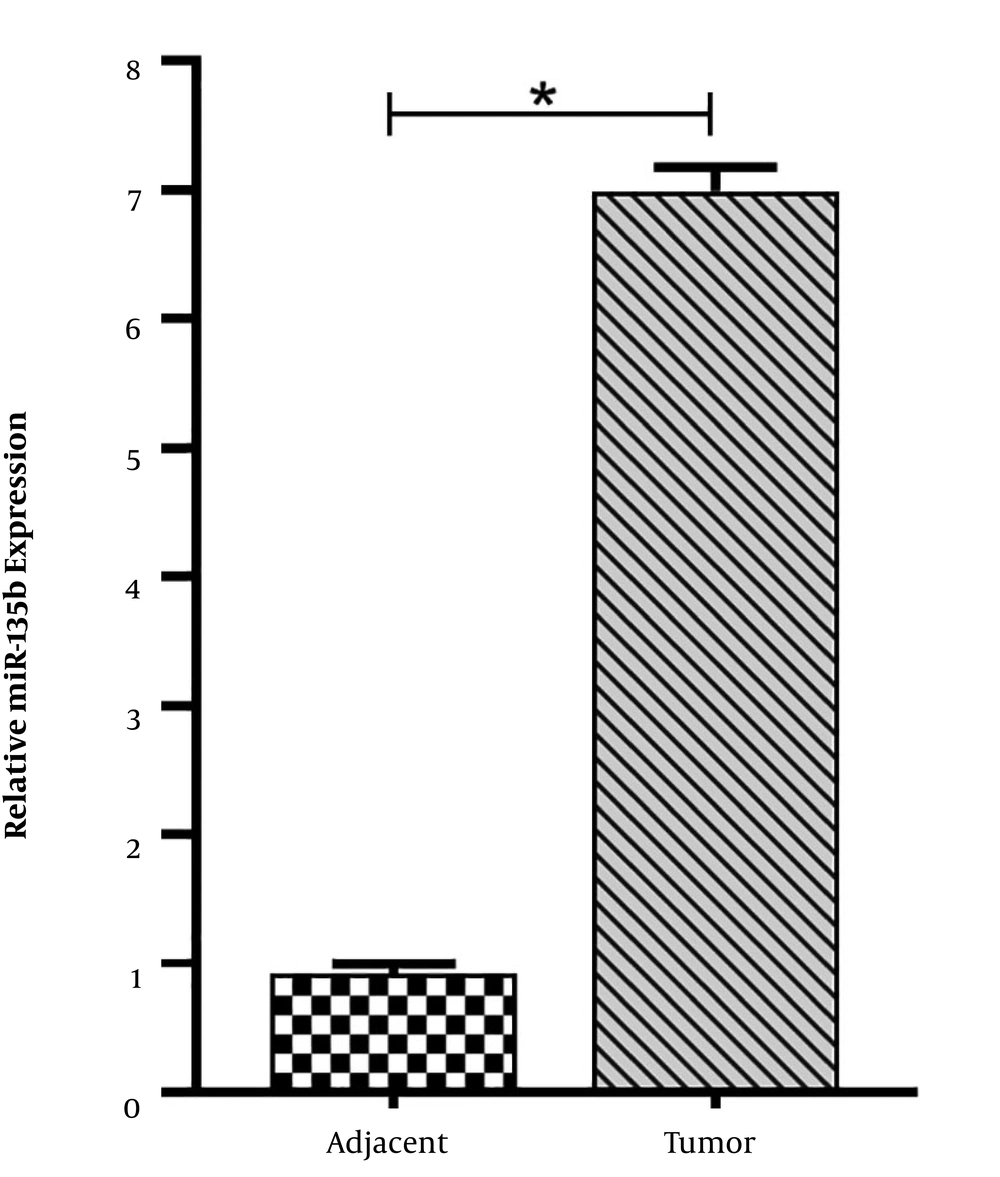
For the assessment of the role of miR-135b in the development of OS, the expression of miR-135b was analyzed in 21 paired OS and adjacent normal bone tissues, using the qRT-PCR method. The results revealed that the expression level of miR-135b was significantly higher in cancerous tissues compared with adjacent normal bone tissues (Figure 1).
4.2. The miR-135b Expression After the Transfection of OS Cell-Line
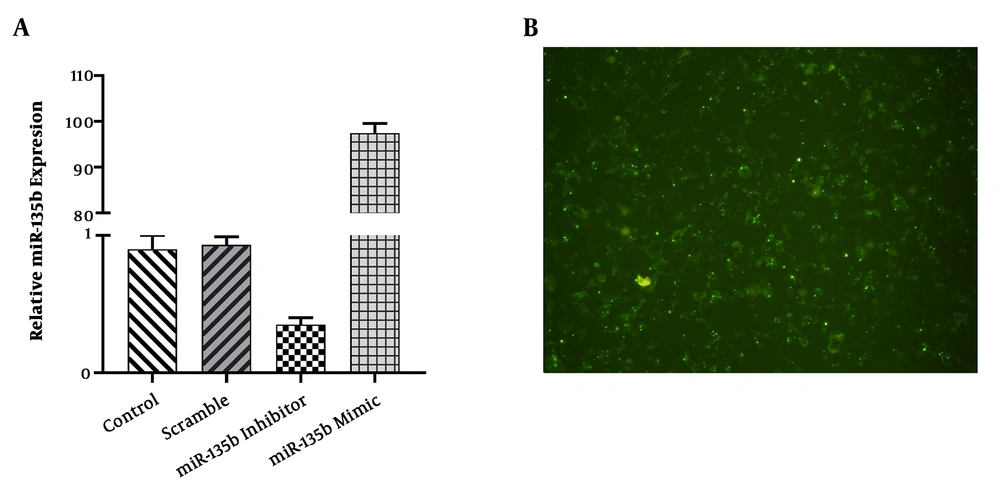
The findings of this study indicated that the expression level of miR-135b was increased by 100 folds in the Saos-2 cell-line transfected with miR-135b mimic, while the expression level of miR-135b was significantly decreased in the Saos-2 cell-line transfected with anti-miR-135b (Figure 2A). The efficiency of the transfection procedure was examined under an Olympus (DP70) microscope (Figure 2B).
4.3. MiR-135b Induces Cell Proliferation in the Saos-2 Cell-Line
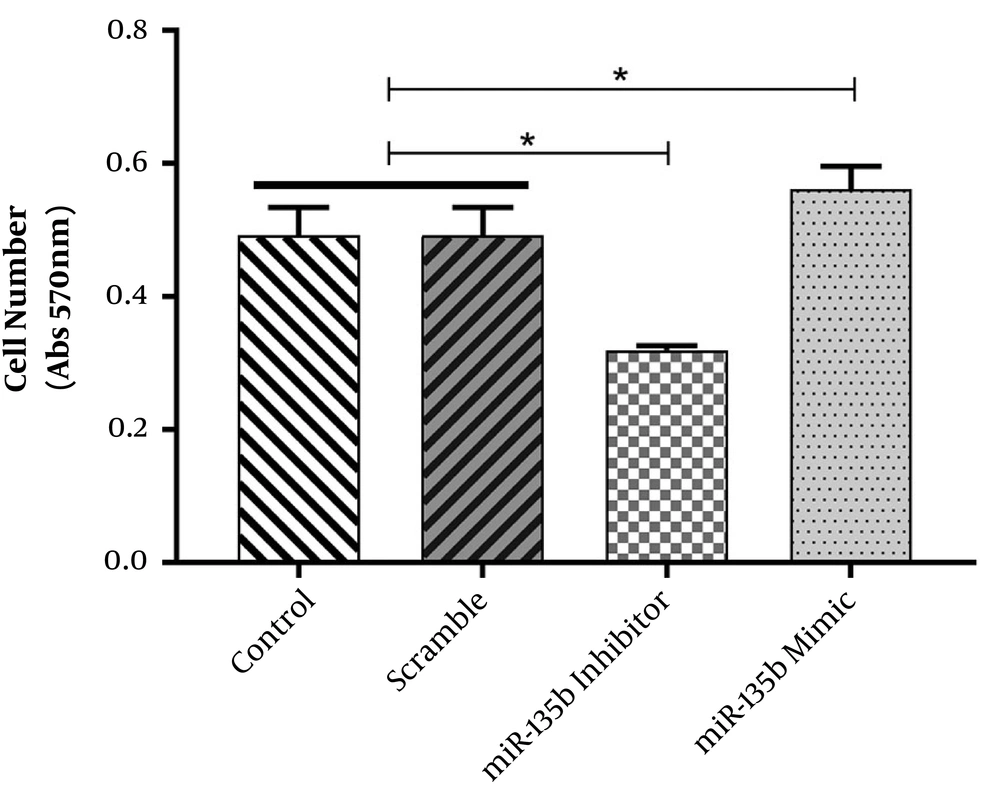
The possible tumorigenic or tumor-suppressive roles of miR-135b in OS were assessed by the in vitro functional analyses, including MTT and cell viability assays. The MTT assay revealed that Saos-2 cells transfected with miR-135b mimic exhibited higher proliferative activity in comparison with cells transfected with scrambled miRNA. Furthermore, the downregulation of miR-135b in Saos-2 cells after transfection with anti-miR-135b was able to decrease cell proliferation (Figure 3). These results indicate that miR-135b exerts an oncogenic function when introduced into these cells.
4.4. Overexpression of miR-135b Accelerates Migration and Invasion of Saos-2 Cells
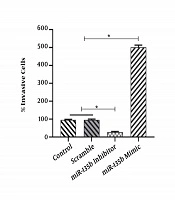
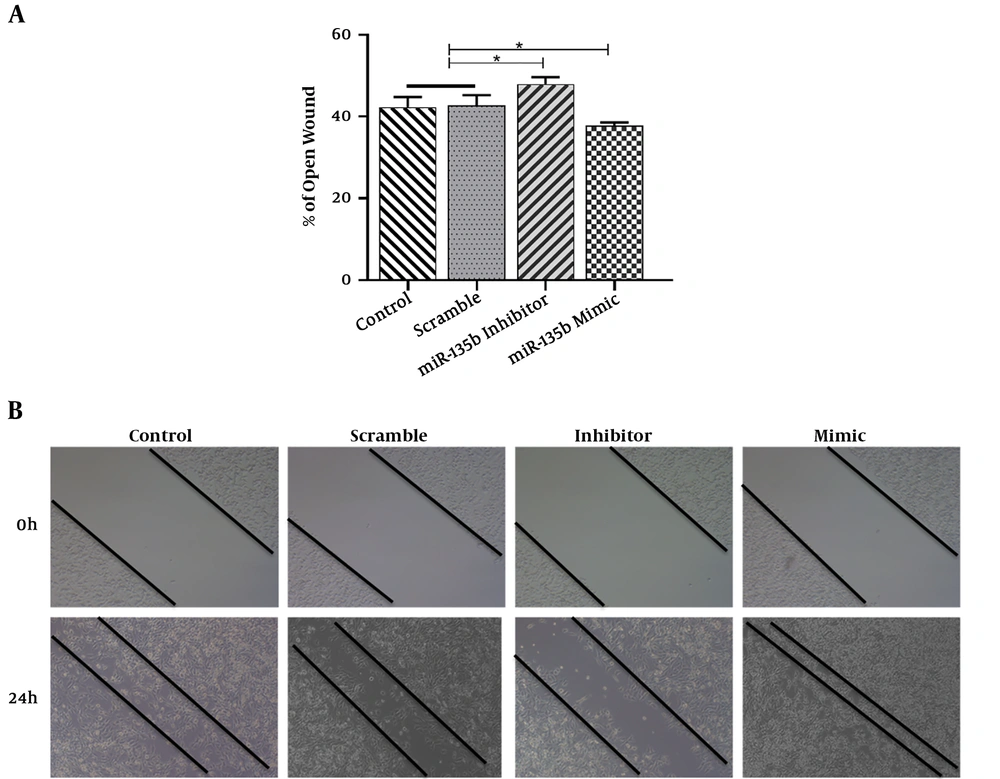
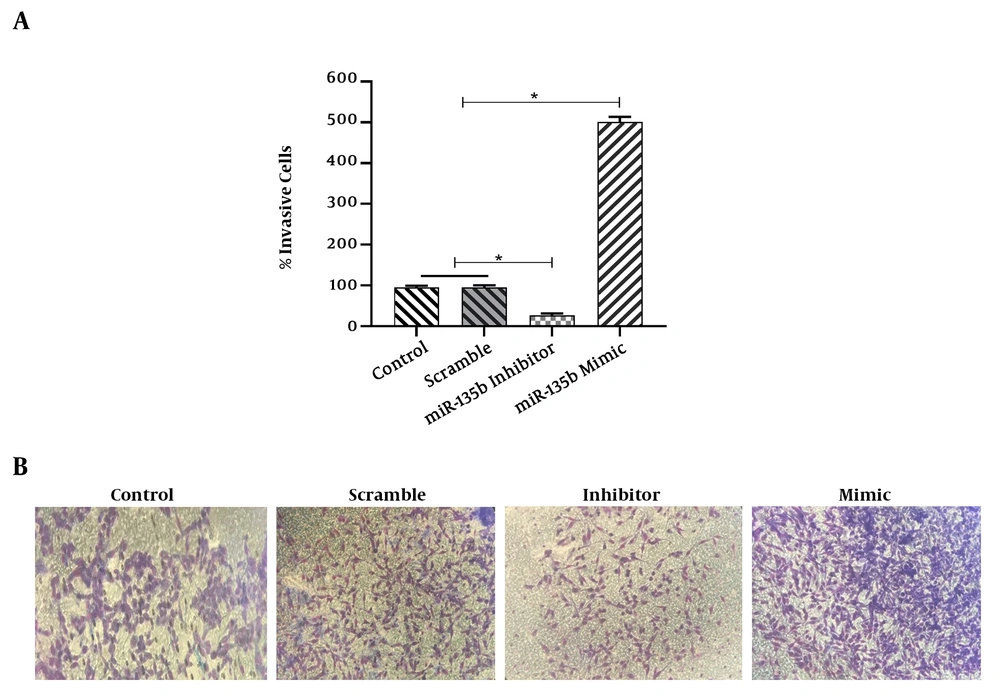
The results of the scratch assay (wound healing assay) showed that the overexpression of miR-135b significantly increased the migration of Saos-2 cells. Cells transfected with the miR-135b mimic have more migratory potential than the cells transfected with control and scrambled miRNAs. Of note, the miR-135b inhibitor markedly decreased the cell migration of Saos-2 cells at 24 hours of the transfection process (Figure 4A). The distance of cell migration was calculated to quantitatively determine the migration ability of the cells (Figure 4B). Consistently, the overexpression of miR-135b induced the cell invasion measured by the transwell assay (Figure 5).
4.5. MiR-135b Targets FOXO-1 in Saos-2 Cell-Line
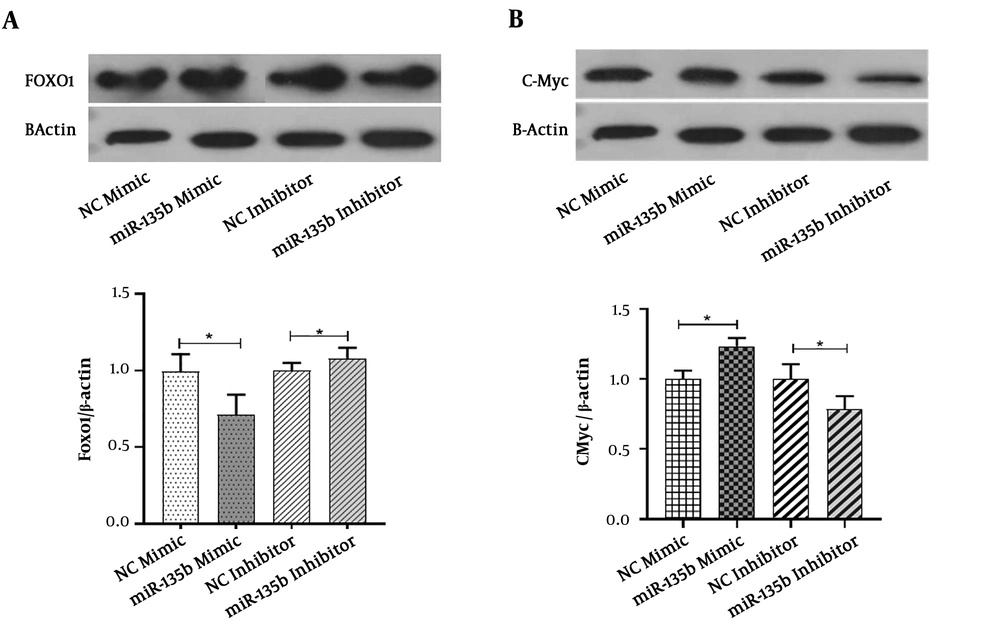
The Saos-2 cell-line was transiently transfected with the miR-135b mimic and inhibitor. The western blot analysis was performed to detect FOXO-1 at the protein level. Compared with the negative control group, the protein level of FOXO-1 was significantly diminished and increased in response to the miR-135b mimic and inhibitor, respectively, when applied for Saos-2 cells (Figure 6A). These results indicated that the FOXO-1 protein contains some binding sites for miR-135b.
MiR-135b targets FOXO-1 and c-Myc in OS cells. A, The protein expression of FOXO-1 in Saos-2 cells transfected with 20 nmol of the miR-135b mimic and inhibitor after 48 hours of the transfection by the western blot analysis; B, the protein expression of c-Myc in Saos-2 cells transfected with 20 nmol of the miR-135b mimic and inhibitor, after 48 hours of the transfection process by the western blot analysis (NC: negative control; *, P < 0.05).
4.6. MiR-135b Induces c-Myc Expression in Saos-2 Cell-Line
The Saos-2 cell-line was transiently transfected with the miR-135b mimic and inhibitor. The findings of the present study demonstrated that in comparison with the negative control group, the protein level of c-Myc was significantly increased and decreased in response to the miR-135b mimic and inhibitor, respectively (Figure 6B). These results showed that the c-Myc protein is upregulated in response to the introduction of miR-135b into the cells.
5. Discussion
The dysregulation of miRNAs has been documented in a wide range of malignancies (14, 16, 33-35). The pattern of alterations in the expression levels of miRNAs is widely used in cancerous tissues for diagnostic and prognostic purposes, as well as finding novel biomarkers for the early screening of patients with cancer (36).
Herein, in line with previous studies, we showed that the expression of miR-135b is aberrantly upregulated in primary tumors of patients with OS compared with the adjacent normal tissues (14, 16, 35, 36). The change in the expression of miR-135b affects some molecular pathways involved in the onset and progression of neoplasia (4). The proliferation, invasion, and metastasis are 3 cellular events that are critical for cancer progression. This research indicated that miR-135b plays a decisive role in the proliferation, invasion, and metastasis of cancer cells. We found that the ectopic expression of miR-135b promoted cell proliferation, invasion, and metastasis when assessed in vitro. In contrast, a significant reduction was detected in cell proliferation, invasion, and metastasis in the presence of endogenous miR-135b inhibitor. Hence, it can be inferred that miR-135b can enhance cell proliferation, invasion, and metastasis in OS cells. Consistently, Lin et al. demonstrated that the expression of miR-135b increases cell invasiveness and metastasis (11, 19).
In the current study, the results have shown that the function of miR-135b is mechanistically dependent on the regulation of the gene expression of FOXO-1 and c-Myc. In detail, we found that the transfection of OS cells with miR-135b mimic suppressed FOXO-1 expression, which in turn led to the induction of proliferation, invasion, and metastasis of the cancer cells. FOXO-1, one of the FOXO family members of the transcription factors, plays a critical role in the proliferation and causes the repression of the cell cycle (3, 37-39) and, more importantly, it is involved in osteocyte survival (3). It has been suggested that the family members of FOXO are oncogenic regulators (40) or tumor-suppressors in a variety of cancers (39). The bioinformatics analysis showed that FOXO-1 is a direct target for miR-135b in OS (14). There is tantalizing evidence showing that miR-135b is one of the potential miRNAs that can downregulate FOXO-1 and promote cancer cell proliferation (41). In parallel, Guan H et al. (3) indicated that FOXO-1 inhibition caused an increase in cell proliferation.
For the determination of the precise role of miR-135b, we evaluated the expression of c-Myc along with FOXO-1. The results of this study showed that the upregulation and downregulation of this miRNA are associated with an increase and a decrease in the protein level of c-Myc in OS cells, respectively.
The mechanistic connection of the genetic and epigenetic alterations with c-Myc activation has been addressed in numerous cancers (2). It is well-established that the c-Myc protein is a multifunctional transcription factor that plays a pivotal role in regulating cellular processes such as cell migration and proliferation (42). Hence, its upregulation can increase the rate of these cellular phenomena (42). Miller et al. (43) demonstrated that miRNAs are potential regulators of c-Myc that they can post-translationally induce the overexpression of this gene. The overexpression of c-Myc has been demonstrated in several types of cancers, as well as its contribution to cancer cell cycle progression, invasion, migration, and metastasis (25). The aberrant expression of c-Myc has been implicated in many neoplasms (44, 45). The uncontrolled expression of c-Myc provides sufficient energy and substrates for the process of cell proliferation (42, 46). Moreover, its activation is associated with the progression of some tumors and poor prognosis (3, 45, 47).
5.1. Conclusions
In conclusion, it seems that miRNA-135b is involved in growth and the spread of OS in a mechanism dependent on modulating the expression of both c-Myc and FOXO-1. Although the results of this investigation, along with others, could partly introduce miRNA-135b as a possible player in OS pathogenesis, more studies are necessary to clarify the role of miRNAs in this regard.